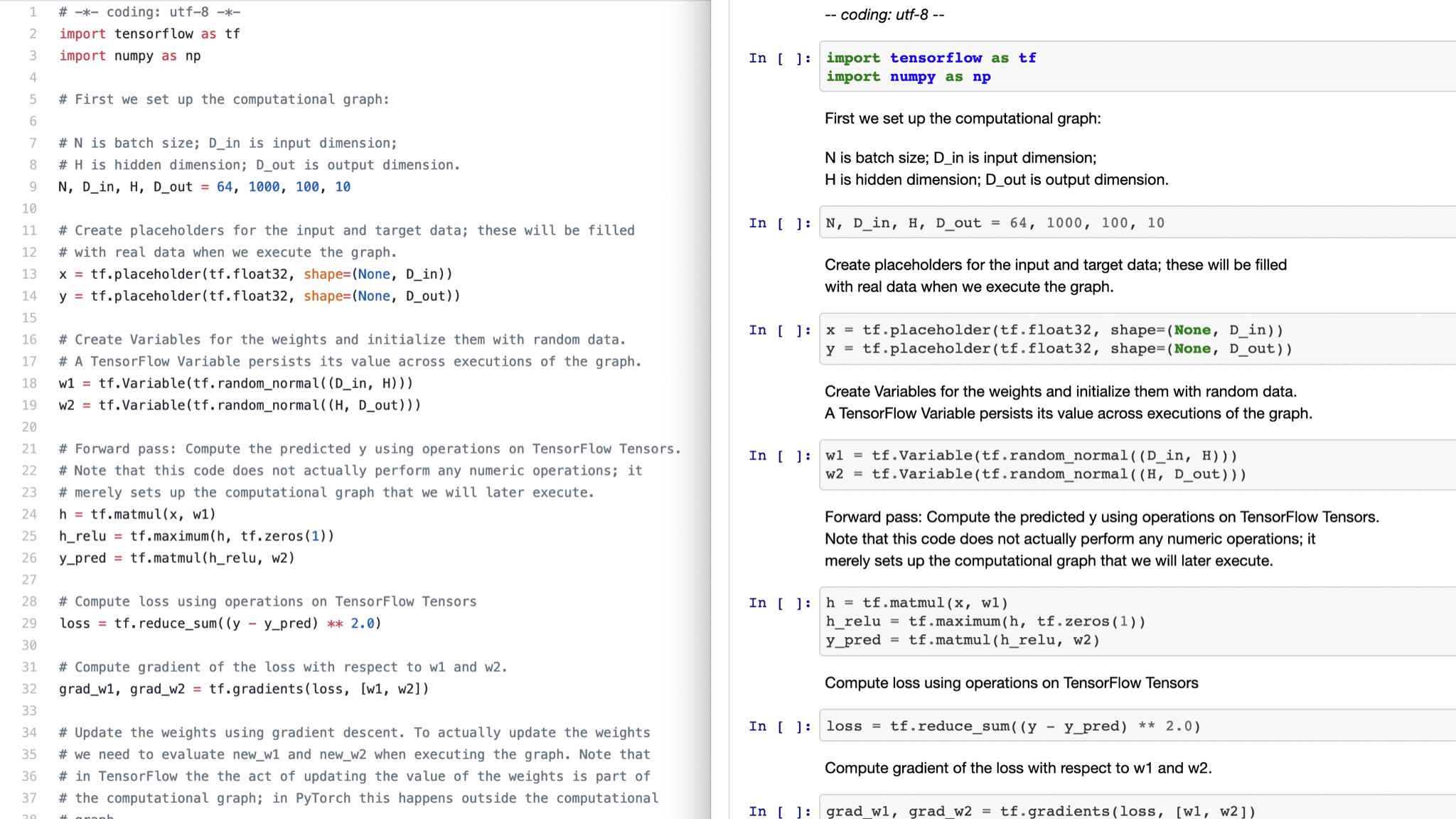
IPython Notebook comes with nbconvert, which can export notebooks to other formats. But how do I convert text in the opposite direction? I ask because I already have materials, and a good workflow, in a different format, but I would like to take advantage of Notebook's interactive environment.
A likely solution: A notebook can be created by importing a .py file, and the documentation states that when nbconvert exports a notebook as a python script, it embeds directives in comments that can be used to recreate the notebook. But the information comes with a disclaimer about the limitations of this method, and the accepted format is not documented anywhere that I could find. (A sample is shown, oddly enough, in the section describing notebook's JSON format). Can anyone provide more information, or a better alternative?
Edit (1 March 2016): The accepted answer no longer works, because for some reason this input format is not supported by version 4 of the Notebook API. I have added a self-answer showing how to import a notebook with the current (v4) API. (I am not un-accepting the current answer, since it solved my problem at the time and pointed me to the resources I used in my self-answer.)