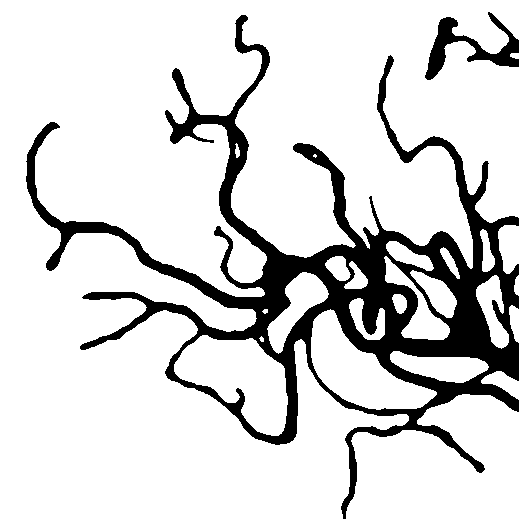
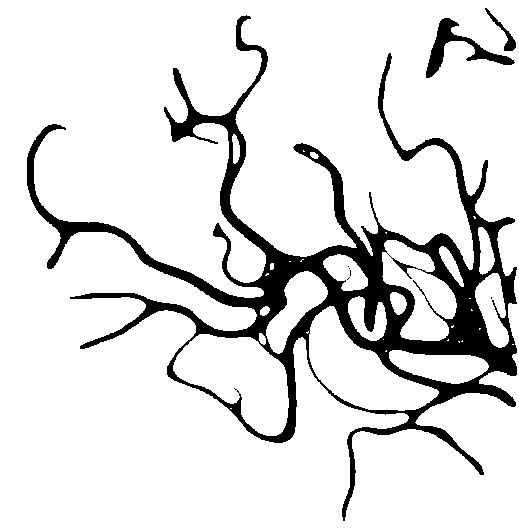
i made some modifications on @dhanushka 's answer for another question and get these images.
Sorry it is C++ code but maybe you will convert it to Python.
![enter image description here]()
You can change the parameters below to get different results.
// contour smoothing parameters for gaussian filter
int filterRadius = 10; // you can try to change this value
int filterSize = 2 * filterRadius + 1;
double sigma = 20; // you can try to change this value
![enter image description here]()
#include <opencv2/highgui.hpp>
#include <opencv2/imgproc.hpp>
#include <iostream>
using namespace cv;
using namespace std;
int main( int argc, const char** argv )
{
Mat im = imread(argv[1], 0);
Mat cont = ~im;
Mat original = Mat::zeros(im.rows, im.cols, CV_8UC3);
Mat smoothed = Mat(im.rows, im.cols, CV_8UC3, Scalar(255,255,255));
// contour smoothing parameters for gaussian filter
int filterRadius = 5;
int filterSize = 2 * filterRadius + 1;
double sigma = 10;
vector<vector<Point> > contours;
vector<Vec4i> hierarchy;
// find contours and store all contour points
findContours(cont, contours, hierarchy, CV_RETR_CCOMP, CV_CHAIN_APPROX_NONE, Point(0, 0));
for(size_t j = 0; j < contours.size(); j++)
{
// extract x and y coordinates of points. we'll consider these as 1-D signals
// add circular padding to 1-D signals
size_t len = contours[j].size() + 2 * filterRadius;
size_t idx = (contours[j].size() - filterRadius);
vector<float> x, y;
for (size_t i = 0; i < len; i++)
{
x.push_back(contours[j][(idx + i) % contours[j].size()].x);
y.push_back(contours[j][(idx + i) % contours[j].size()].y);
}
// filter 1-D signals
vector<float> xFilt, yFilt;
GaussianBlur(x, xFilt, Size(filterSize, filterSize), sigma, sigma);
GaussianBlur(y, yFilt, Size(filterSize, filterSize), sigma, sigma);
// build smoothed contour
vector<vector<Point> > smoothContours;
vector<Point> smooth;
for (size_t i = filterRadius; i < contours[j].size() + filterRadius; i++)
{
smooth.push_back(Point(xFilt[i], yFilt[i]));
}
smoothContours.push_back(smooth);
Scalar color;
if(hierarchy[j][3] < 0 )
{
color = Scalar(0,0,0);
}
else
{
color = Scalar(255,255,255);
}
drawContours(smoothed, smoothContours, 0, color, -1);
}
imshow( "result", smoothed );
waitKey(0);
}