I'm new to Granger Causality and would appreciate any advice on understanding/interpreting the results of the python statsmodels output. I've constructed two data sets (sine functions shifted in time with noise added)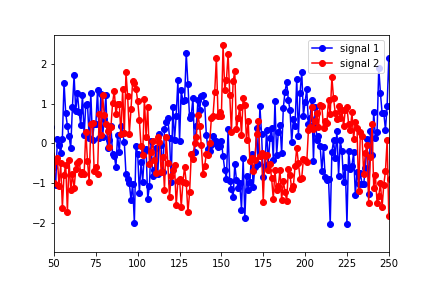
and put them in a "data" matrix with signal 1 as the first column and signal 2 as the second. I then ran the tests using:
granger_test_result = sm.tsa.stattools.grangercausalitytests(data, maxlag=40, verbose=True)`
The results showed that the optimal lag (in terms of the highest F test value) were for a lag of 1.
Granger Causality
('number of lags (no zero)', 1)
ssr based F test: F=96.6366 , p=0.0000 , df_denom=995, df_num=1
ssr based chi2 test: chi2=96.9280 , p=0.0000 , df=1
likelihood ratio test: chi2=92.5052 , p=0.0000 , df=1
parameter F test: F=96.6366 , p=0.0000 , df_denom=995, df_num=1
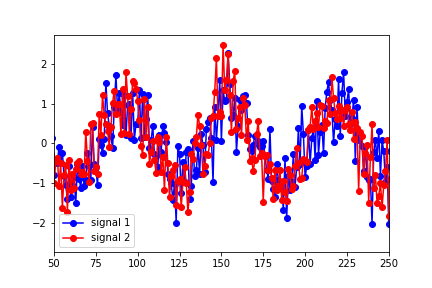
However, the lag that seems to best describe the optimal overlap of the data is around 25 (in the figure below, signal 1 has been shifted to the right by 25 points):
Granger Causality
('number of lags (no zero)', 25)
ssr based F test: F=4.1891 , p=0.0000 , df_denom=923, df_num=25
ssr based chi2 test: chi2=110.5149, p=0.0000 , df=25
likelihood ratio test: chi2=104.6823, p=0.0000 , df=25
parameter F test: F=4.1891 , p=0.0000 , df_denom=923, df_num=25
I'm clearly misinterpreting something here. Why wouldn't the predicted lag match up with the shift in the data?
Also, can anyone explain to me why the p-values are so small as to be negligible for most lag values? They only begin to show up as non-zero for lags greater than 30.
Thanks for any help you can give.
