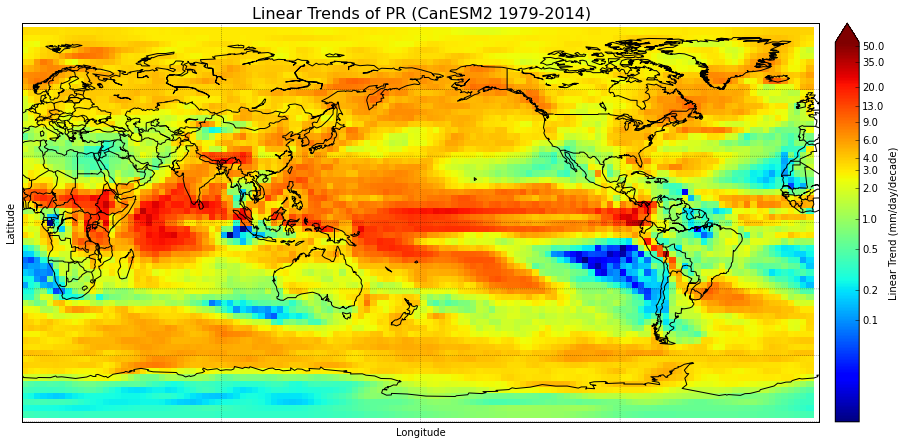
I have a basemap of the world, and it's filled with data (lintrends_mean) using pcolormesh. Because the data has relatively large grid boxes, I'd like to smooth the plot. However, I can't figure out how to do this. Setting shading='gouraud' in the plotting function blurs the edges of the grid boxes, but I'd like something nicer-looking than that since the data still appears blotchy.
There was a similar question asked here with an answer given, but I don't understand the answer, particularly where the "newdepth" comes from. I can't comment on it for clarification either since I'm short on reputation. interpolation with matplotlib pcolor
#Set cmap properties
bounds = np.array([0.1,0.2,0.5,1,2,3,4,6,9,13,20,35,50])
norm = colors.LogNorm(vmin=0.01,vmax=55) #creates logarithmic scale
#cmap.set_under('#000099') # I want to use this- edit in Paint
cmap.set_over('#660000') # everything above range of colormap
fig = plt.figure(figsize=(15.,10.)) #create figure & size
m = Basemap(projection='cyl',llcrnrlat=-90,urcrnrlat=90,llcrnrlon=0,urcrnrlon=360.,lon_0=180.,resolution='c') #create basemap & specify data area & res
m.drawcoastlines(linewidth=1)
m.drawcountries(linewidth=1)
m.drawparallels(np.arange(-90,90,30.),linewidth=0.3)
m.drawmeridians(np.arange(-180.,180.,90.),linewidth=0.3)
meshlon,meshlat = np.meshgrid(lon,lat) #meshgrid turns lats & lons into 2D arrays
x,y = m(meshlon,meshlat) #assign 2D arrays to new variables
trend = m.pcolormesh(x,y,lintrends_mean,cmap=plt.get_cmap('jet'),norm=norm) #plot the data & specify colormap & color range
cbar=m.colorbar(trend,size="3%", label='Linear Trend (mm/day/decade)',ticks=bounds,extend="max")
cbar.set_ticklabels(bounds)
plt.title('Linear Trends of PR (CanESM2 1979-2014)',fontsize=16)
plt.xlabel('Longitude',fontsize=10)
plt.ylabel('Latitude',fontsize=10)
plt.show()