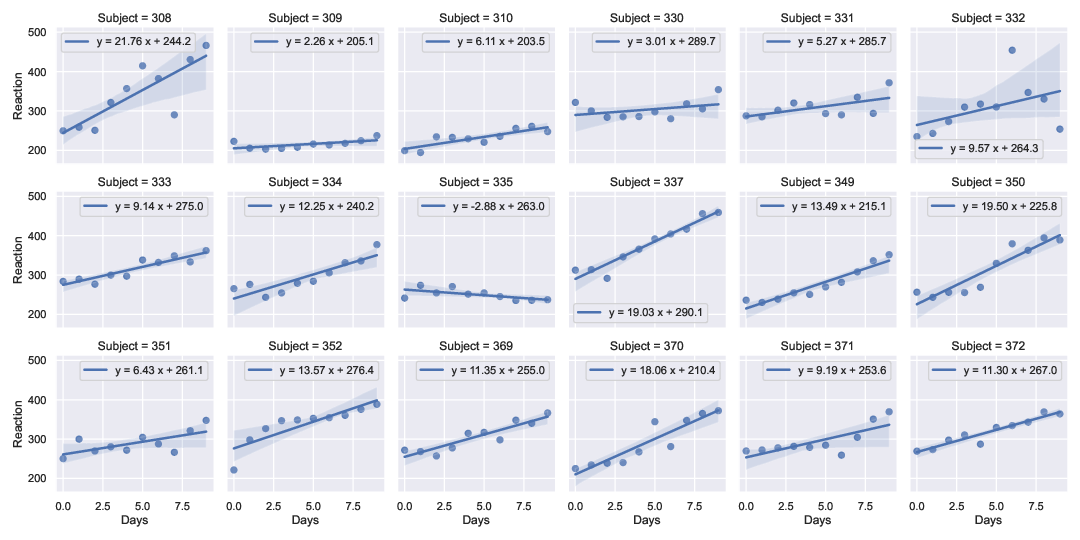
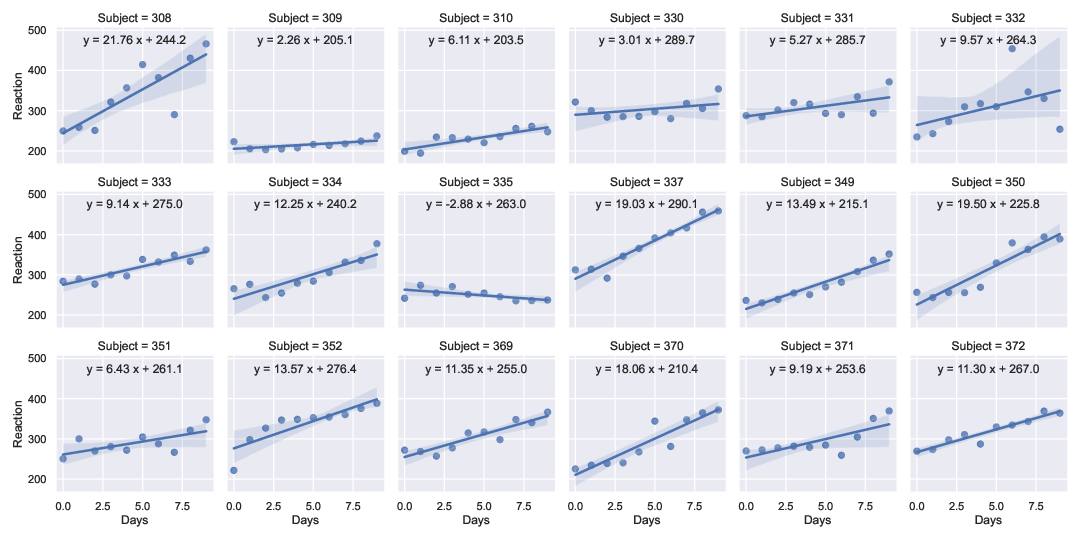
I extended the solution by @RMS to work for a multi-panel lmplot example (using data from a sleep-deprivation study (Belenky et. al., J Sleep Res 2003) available in pydataset). This allows one to have axis-specific legends/labels without having to use, e.g., regplot and plt.subplots.
Edit: Added second method using the map_dataframe() method from FacetGrid(), as suggested in the answer by Marcos here.
import numpy as np
import scipy as sp
import pandas as pd
import seaborn as sns
import pydataset as pds
import matplotlib.pyplot as plt
# use seaborn theme
sns.set_theme(color_codes=True)
# Load data from sleep deprivation study (Belenky et al, J Sleep Res 2003)
# ['Reaction', 'Days', 'Subject'] = [reaction time (ms), deprivation time, Subj. No.]
df = pds.data("sleepstudy")
# convert integer label to string
df['Subject'] = df['Subject'].apply(str)
# perform linear regressions outside of seaborn to get parameters
subjects = np.unique(df['Subject'].to_numpy())
fit_str = []
for s in subjects:
ddf = df[df['Subject'] == s]
m, b, r_value, p_value, std_err = \
sp.stats.linregress(ddf['Days'],ddf['Reaction'])
fs = f"y = {m:.2f} x + {b:.1f}"
fit_str.append(fs)
method_one = False
method_two = True
if method_one:
# Access legend on each axis to write equation
#
# Create 18 panel lmplot with seaborn
g = sns.lmplot(x="Days", y="Reaction", col="Subject",
col_wrap=6, height=2.5, data=df,
line_kws={'label':"Linear Reg"}, legend=True)
# write string with fit result into legend string of each axis
axes = g.axes # 18 element list of axes objects
i=0
for ax in axes:
ax.legend() # create legend on axis
leg = ax.get_legend()
leg_labels = leg.get_texts()
leg_labels[0].set_text(fit_str[i])
i += 1
elif method_two:
# use the .map_dataframe () method from FacetGrid() to annotate plot
# https://stackoverflow.com/questions/25579227 (answer by @Marcos)
#
# Create 18 panel lmplot with seaborn
g = sns.lmplot(x="Days", y="Reaction", col="Subject",
col_wrap=6, height=2.5, data=df)
def annotate(data, **kws):
m, b, r_value, p_value, std_err = \
sp.stats.linregress(data['Days'],data['Reaction'])
ax = plt.gca()
ax.text(0.5, 0.9, f"y = {m:.2f} x + {b:.1f}",
horizontalalignment='center',
verticalalignment='center',
transform=ax.transAxes)
g.map_dataframe(annotate)
# write figure to pdf
plt.savefig("sleepstudy_data_w-fits.pdf")
Output (Method 1):
![enter image description here]()
Output (Method 2):
![enter image description here]()
Update 2022-05-11: Unrelated to the plotting techniques, it turns out that this interpretation of the data (and that provided, e.g., in the original R repository) is incorrect. See the reported issue here. Fits should be done to days 2-9, corresponding to zero to seven days of sleep deprivation (3h sleep per night). The first three data points correspond to training and baseline days (all with 8h sleep per night).