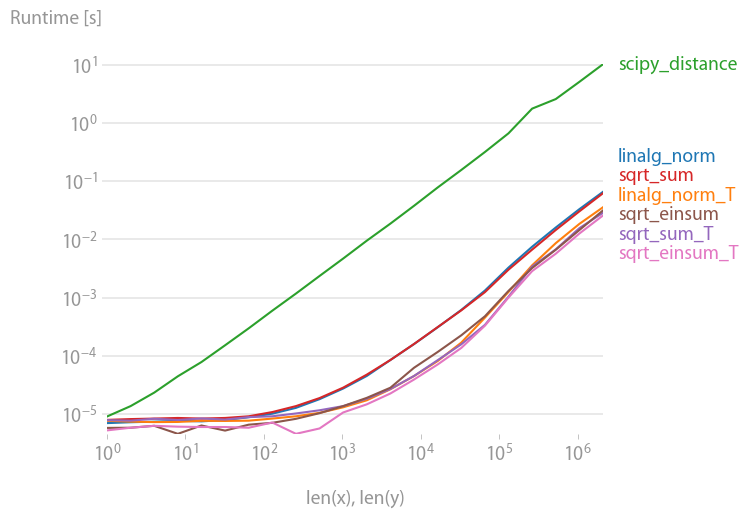
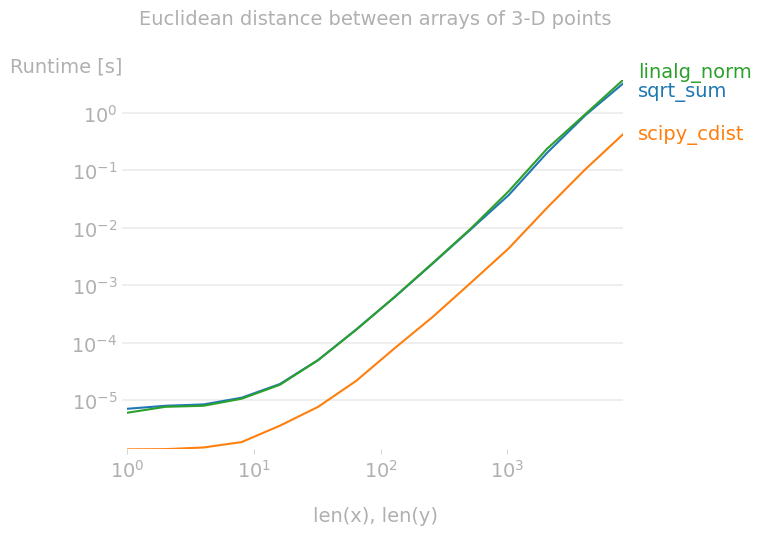
I want to expound on the simple answer with various performance notes. np.linalg.norm will do perhaps more than you need:
dist = numpy.linalg.norm(a-b)
Firstly - this function is designed to work over a list and return all of the values, e.g. to compare the distance from pA to the set of points sP:
sP = set(points)
pA = point
distances = np.linalg.norm(sP - pA, ord=2, axis=1.) # 'distances' is a list
Remember several things:
- Python function calls are expensive.
- [Regular] Python doesn't cache name lookups.
So
def distance(pointA, pointB):
dist = np.linalg.norm(pointA - pointB)
return dist
isn't as innocent as it looks.
>>> dis.dis(distance)
2 0 LOAD_GLOBAL 0 (np)
2 LOAD_ATTR 1 (linalg)
4 LOAD_ATTR 2 (norm)
6 LOAD_FAST 0 (pointA)
8 LOAD_FAST 1 (pointB)
10 BINARY_SUBTRACT
12 CALL_FUNCTION 1
14 STORE_FAST 2 (dist)
3 16 LOAD_FAST 2 (dist)
18 RETURN_VALUE
Firstly - every time we call it, we have to do a global lookup for "np", a scoped lookup for "linalg" and a scoped lookup for "norm", and the overhead of merely calling the function can equate to dozens of python instructions.
Lastly, we wasted two operations on to store the result and reload it for return...
First pass at improvement: make the lookup faster, skip the store
def distance(pointA, pointB, _norm=np.linalg.norm):
return _norm(pointA - pointB)
We get the far more streamlined:
>>> dis.dis(distance)
2 0 LOAD_FAST 2 (_norm)
2 LOAD_FAST 0 (pointA)
4 LOAD_FAST 1 (pointB)
6 BINARY_SUBTRACT
8 CALL_FUNCTION 1
10 RETURN_VALUE
The function call overhead still amounts to some work, though. And you'll want to do benchmarks to determine whether you might be better doing the math yourself:
def distance(pointA, pointB):
return (
((pointA.x - pointB.x) ** 2) +
((pointA.y - pointB.y) ** 2) +
((pointA.z - pointB.z) ** 2)
) ** 0.5 # fast sqrt
On some platforms, **0.5 is faster than math.sqrt. Your mileage may vary.
**** Advanced performance notes.
Why are you calculating distance? If the sole purpose is to display it,
print("The target is %.2fm away" % (distance(a, b)))
move along. But if you're comparing distances, doing range checks, etc., I'd like to add some useful performance observations.
Let’s take two cases: sorting by distance or culling a list to items that meet a range constraint.
# Ultra naive implementations. Hold onto your hat.
def sort_things_by_distance(origin, things):
return things.sort(key=lambda thing: distance(origin, thing))
def in_range(origin, range, things):
things_in_range = []
for thing in things:
if distance(origin, thing) <= range:
things_in_range.append(thing)
The first thing we need to remember is that we are using Pythagoras to calculate the distance (dist = sqrt(x^2 + y^2 + z^2)) so we're making a lot of sqrt calls. Math 101:
dist = root ( x^2 + y^2 + z^2 )
:.
dist^2 = x^2 + y^2 + z^2
and
sq(N) < sq(M) iff M > N
and
sq(N) > sq(M) iff N > M
and
sq(N) = sq(M) iff N == M
In short: until we actually require the distance in a unit of X rather than X^2, we can eliminate the hardest part of the calculations.
# Still naive, but much faster.
def distance_sq(left, right):
""" Returns the square of the distance between left and right. """
return (
((left.x - right.x) ** 2) +
((left.y - right.y) ** 2) +
((left.z - right.z) ** 2)
)
def sort_things_by_distance(origin, things):
return things.sort(key=lambda thing: distance_sq(origin, thing))
def in_range(origin, range, things):
things_in_range = []
# Remember that sqrt(N)**2 == N, so if we square
# range, we don't need to root the distances.
range_sq = range**2
for thing in things:
if distance_sq(origin, thing) <= range_sq:
things_in_range.append(thing)
Great, both functions no-longer do any expensive square roots. That'll be much faster, but before you go further, check yourself: why did sort_things_by_distance need a "naive" disclaimer both times above? Answer at the very bottom (*a1).
We can improve in_range by converting it to a generator:
def in_range(origin, range, things):
range_sq = range**2
yield from (thing for thing in things
if distance_sq(origin, thing) <= range_sq)
This especially has benefits if you are doing something like:
if any(in_range(origin, max_dist, things)):
...
But if the very next thing you are going to do requires a distance,
for nearby in in_range(origin, walking_distance, hotdog_stands):
print("%s %.2fm" % (nearby.name, distance(origin, nearby)))
consider yielding tuples:
def in_range_with_dist_sq(origin, range, things):
range_sq = range**2
for thing in things:
dist_sq = distance_sq(origin, thing)
if dist_sq <= range_sq: yield (thing, dist_sq)
This can be especially useful if you might chain range checks ('find things that are near X and within Nm of Y', since you don't have to calculate the distance again).
But what about if we're searching a really large list of things and we anticipate a lot of them not being worth consideration?
There is actually a very simple optimization:
def in_range_all_the_things(origin, range, things):
range_sq = range**2
for thing in things:
dist_sq = (origin.x - thing.x) ** 2
if dist_sq <= range_sq:
dist_sq += (origin.y - thing.y) ** 2
if dist_sq <= range_sq:
dist_sq += (origin.z - thing.z) ** 2
if dist_sq <= range_sq:
yield thing
Whether this is useful will depend on the size of 'things'.
def in_range_all_the_things(origin, range, things):
range_sq = range**2
if len(things) >= 4096:
for thing in things:
dist_sq = (origin.x - thing.x) ** 2
if dist_sq <= range_sq:
dist_sq += (origin.y - thing.y) ** 2
if dist_sq <= range_sq:
dist_sq += (origin.z - thing.z) ** 2
if dist_sq <= range_sq:
yield thing
elif len(things) > 32:
for things in things:
dist_sq = (origin.x - thing.x) ** 2
if dist_sq <= range_sq:
dist_sq += (origin.y - thing.y) ** 2 + (origin.z - thing.z) ** 2
if dist_sq <= range_sq:
yield thing
else:
... just calculate distance and range-check it ...
And again, consider yielding the dist_sq. Our hotdog example then becomes:
# Chaining generators
info = in_range_with_dist_sq(origin, walking_distance, hotdog_stands)
info = (stand, dist_sq**0.5 for stand, dist_sq in info)
for stand, dist in info:
print("%s %.2fm" % (stand, dist))
(*a1: sort_things_by_distance's sort key calls distance_sq for every single item, and that innocent looking key is a lambda, which is a second function that has to be invoked...)