I'd like to explain an O(V * (V + E)) solution to this problem, which on sparse graphs is significantly more efficient than O(V^3) Floyd-Warshall.
First, let's fix the starting node of the cycle to be node S. Then, let's compute
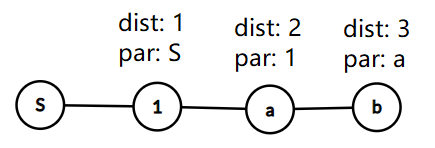
dist[i] = the shortest distance from node S to node i
par[i] = the parent of node i; any node j where dist[i] = dist[j] + 1
in O(V + E) time using a simple BFS.
Now, the key idea is to find, for each edge, the shortest cycle from S that passes through that edge. Then, because any cycle must pass through some edge of the graph, the shortest of all shortest cycles that pass through a given edge will yield the shortest cycle starting at S.
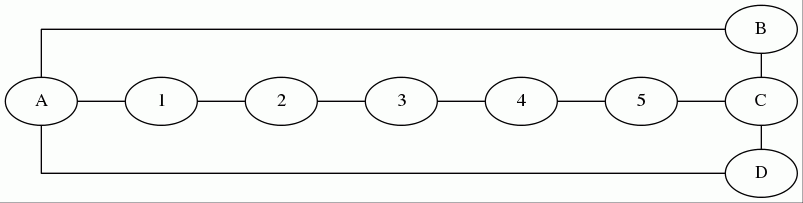
But how do we find the shortest cycle from S that passes through a given undirected edge (a, b)? The shortest length is NOT just dist[a] + dist[b] + 1 (from the cycle S -> a -> b -> S), because the paths S -> a and b -> S may intersect. Consider the graph below:
![enter image description here]()
Here, if we used dist[a] + dist[b] + 1, then we'd report a cycle of length 2 + 3 + 1 = 6, when in reality no cycle exists at all because the path from S -> a is a prefix of the path from S -> b.
So, we need to ensure that for (a, b), the path S -> a isn't a prefix of S -> b (or vice versa). Observe that this simply equivalent to checking if parent_of[a] != b && parent_of[b] != a. So as long as that condition is true, then we can use dist[a] + dist[b] + 1.
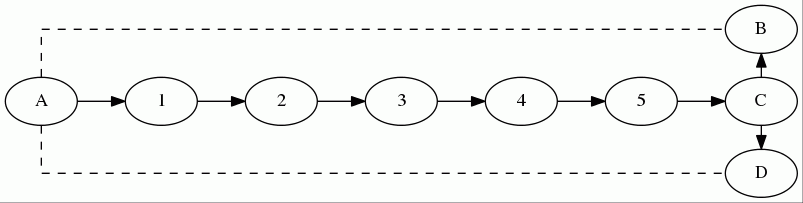
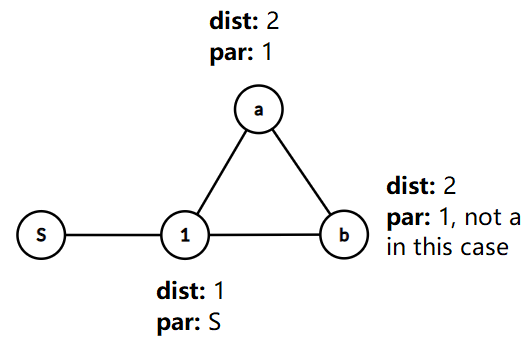
But, you might ask, what if the two paths S -> a and S -> b aren't prefixes of each other, but they still intersect, like below?
![enter image description here]()
Here, because par[b] equals 1 and not a, neither of S->a and S->b are prefixes of the other, so we would report a cycle of length dist[a] + dist[b] + 1 = 2 + 2 + 1 = 5 (representing the cycle S -> 1 -> a -> b -> 1 -> S)! Clearly, this is not a simple cycle, and so shouldn't be counted. However, observe that we do not actually need to change the algorithm to correct this, because (a) we never reports a cycle when one doesn't exist (For instance, although S -> 1 -> a -> b -> 1 -> S isn't a simple cycle, it contains the simple cycle 1 -> a -> b -> 1. Essentially, whenever S->a and S->b share a prefix, then there is an existing cycle that can be found by removing their shared prefix), and (b) this only ever reports cycles longer than the minimum-length cycle, meaning we won't ever get a wrong answer (we won't ever get a cycle shorter than the shortest cycle). And lastly, we will always find the shortest cycle whenever we test a starting node that lies within that cycle, so we will always get the correct answer.
So, to recap:
- From each starting node, we will use a BFS to find the shortest cycle which passes through it.
- Specifically, use the BFS to calculate
dist[i] and par[i].
- Afterwards, enumerate every edge
(a, b), and find the shortest cycle starting from S that passes through that edge.
- As long as
parent_of[a] != b && parent_of[b] != a (meaning neither of the paths S->a and S->b are a prefix of the other, so we don't ever report a cycle where none exists), we have a cycle of length dist[a] + dist[b] + 1.
- Taking the minimum of all
dist[a] + dist[b] + 1 over all edges over all starting nodes, yields the answer.
Time Complexity: O(V(V + E)), since for each of the V starting nodes we run a O(V + E) BFS.
Space Complexity: O(V + E), since we need O(V + E) for the adjacency list and O(V) for the arrays/queues used in the BFS.
Implementation in C++:
int shortest_cycle_length(const vector<vector<int>> &adj) {
int N = adj.size(), INF = N + 1;
int shortest_cycle = INF; // cycle length must be <= N
// Test every possible starting node of the shortest cycle
for (int starting_node = 0; starting_node < N; ++starting_node) {
// Find the shortest cycle that starts (and ends) at starting_node
// BFS, finding for each node its shortest distance from starting_node
// as well as its parent node in the shortest path to it
queue<int> Q;
vector<bool> visited(N, false);
vector<int> dist(N, INF), parent_of(N, -1);
Q.push(starting_node);
visited[starting_node] = true;
dist[starting_node] = 0;
while (!Q.empty()) {
int curr = Q.front();
Q.pop();
for (int next_node : adj[curr]) {
// If we haven't visited next_node, enqueue it (normal BFS)
if (!visited[next_node]) {
visited[next_node] = true;
dist[next_node] = dist[curr] + 1;
parent_of[next_node] = curr;
Q.push(next_node);
}
// Otherwise, we have visited next_node previously.
else if (parent_of[curr] != next_node) {
// ^^ Note that we don't need to check if parent_of[next_node] = curr,
// because that can never happen
// Then there is a cycle from starting_node through curr <=> next_node
// Specifically: starting_node -> curr -> next_node -> starting_node
// Clearly, the shortest length of this cycle is (dist[curr] + dist[next_node] + 1)
shortest_cycle = min(shortest_cycle, dist[curr] + dist[next_node] + 1);
}
}
}
}
// return -1 if no cycle exists, otherwise return shortest_cycle
return (shortest_cycle == INF ? -1 : shortest_cycle);
}
Note that in this implementation, I create new dist and par arrays for each starting node. If you just allocate them once at the beginning and reset them for each new starting node, you'll probably save a bit of time.
Also, notice that instead of iterating over all edges after the BFS is complete and finding the shortest cycle through each edge, I do it during the BFS. When we're at curr_node and wanting to move to next_node, then if next_node has already been visited (so we know it's dist and par values), I find right then and there the shortest cycle through the edge (curr_node, next_node). To do this, we just need to check if parent_of[curr_node] != next_node && parent_of[next_node] != curr_node, but then observe that parent_of[next_node] can never equal curr_node, because otherwise that means next_node could not have been visited previously. As a result, we only need to check if parent_of[curr_node] != next_node; if this condition is satisfied, the minimize the answer with dist[curr_node] + dist[next_node] + 1.