I am in dire need of a classification task example using LibSVM in python. I don't know how the Input should look like and which function is responsible for training and which one for testing Thanks
LIBSVM reads the data from a tuple containing two lists. The first list contains the classes and the second list contains the input data. create simple dataset with two possible classes you also need to specify which kernel you want to use by creating svm_parameter.
>> from libsvm import *
>> prob = svm_problem([1,-1],[[1,0,1],[-1,0,-1]])
>> param = svm_parameter(kernel_type = LINEAR, C = 10)
## training the model
>> m = svm_model(prob, param)
#testing the model
>> m.predict([1, 1, 1])
from svmutil import * instead. See @ShinNoNoir's answer below. –
Centripetal The code examples listed here don't work with LibSVM 3.1, so I've more or less ported the example by mossplix:
from svmutil import *
svm_model.predict = lambda self, x: svm_predict([0], [x], self)[0][0]
prob = svm_problem([1,-1], [[1,0,1], [-1,0,-1]])
param = svm_parameter()
param.kernel_type = LINEAR
param.C = 10
m=svm_train(prob, param)
m.predict([1,1,1])
This example demonstrates a one-class SVM classifier; it's about as simple as possible while still showing the complete LIBSVM workflow.
Step 1: Import NumPy & LIBSVM
import numpy as NP
from svm import *
Step 2: Generate synthetic data: for this example, 500 points within a given boundary (note: quite a few real data sets are are provided on the LIBSVM website)
Data = NP.random.randint(-5, 5, 1000).reshape(500, 2)
Step 3: Now, choose some non-linear decision boundary for a one-class classifier:
rx = [ (x**2 + y**2) < 9 and 1 or 0 for (x, y) in Data ]
Step 4: Next, arbitrarily partition the data w/r/t this decision boundary:
Class I: those that lie on or within an arbitrary circle
Class II: all points outside the decision boundary (circle)
The SVM Model Building begins here; all steps before this one were just to prepare some synthetic data.
Step 5: Construct the problem description by calling svm_problem, passing in the decision boundary function and the data, then bind this result to a variable.
px = svm_problem(rx, Data)
Step 6: Select a kernel function for the non-linear mapping
For this exmaple, i chose RBF (radial basis function) as my kernel function
pm = svm_parameter(kernel_type=RBF)
Step 7: Train the classifier, by calling svm_model, passing in the problem description (px) & kernel (pm)
v = svm_model(px, pm)
Step 8: Finally, test the trained classifier by calling predict on the trained model object ('v')
v.predict([3, 1])
# returns the class label (either '1' or '0')
For the example above, I used version 3.0 of LIBSVM (the current stable release at the time this answer was posted).
Finally, w/r/t the part of your question regarding the choice of kernel function, Support Vector Machines are not specific to a particular kernel function--e.g., i could have chosen a different kernel (gaussian, polynomial, etc.).
LIBSVM includes all of the most commonly used kernel functions--which is a big help because you can see all plausible alternatives and to select one for use in your model, is just a matter of calling svm_parameter and passing in a value for kernel_type (a three-letter abbreviation for the chosen kernel).
Finally, the kernel function you choose for training must match the kernel function used against the testing data.
Traceback (most recent call last): File "<stdin>", line 1, in <module> File "/usr/lib/pymodules/python2.7/svm.py", line 83, in __init__ tmp_xi, tmp_idx = gen_svm_nodearray(xi,isKernel=isKernel) File "/usr/lib/pymodules/python2.7/svm.py", line 51, in gen_svm_nodearray raise TypeError('xi should be a dictionary, list or tuple') TypeError: xi should be a dictionary, list or tuple –
Dire TypeError: __init__() got an unexpected keyword argument 'kernel_type'. –
Soloist LIBSVM reads the data from a tuple containing two lists. The first list contains the classes and the second list contains the input data. create simple dataset with two possible classes you also need to specify which kernel you want to use by creating svm_parameter.
>> from libsvm import *
>> prob = svm_problem([1,-1],[[1,0,1],[-1,0,-1]])
>> param = svm_parameter(kernel_type = LINEAR, C = 10)
## training the model
>> m = svm_model(prob, param)
#testing the model
>> m.predict([1, 1, 1])
from svmutil import * instead. See @ShinNoNoir's answer below. –
Centripetal You might consider using
http://scikit-learn.sourceforge.net/
That has a great python binding of libsvm and should be easy to install
Adding to @shinNoNoir :
param.kernel_type represents the type of kernel function you want to use, 0: Linear 1: polynomial 2: RBF 3: Sigmoid
Also have in mind that, svm_problem(y,x) : here y is the class labels and x is the class instances and x and y can only be lists,tuples and dictionaries.(no numpy array)
SVM via SciKit-learn:
from sklearn.svm import SVC
X = [[0, 0], [1, 1]]
y = [0, 1]
model = SVC().fit(X, y)
tests = [[0.,0.], [0.49,0.49], [0.5,0.5], [2., 2.]]
print(model.predict(tests))
# prints [0 0 1 1]
For more details here: http://scikit-learn.org/stable/modules/svm.html#svm
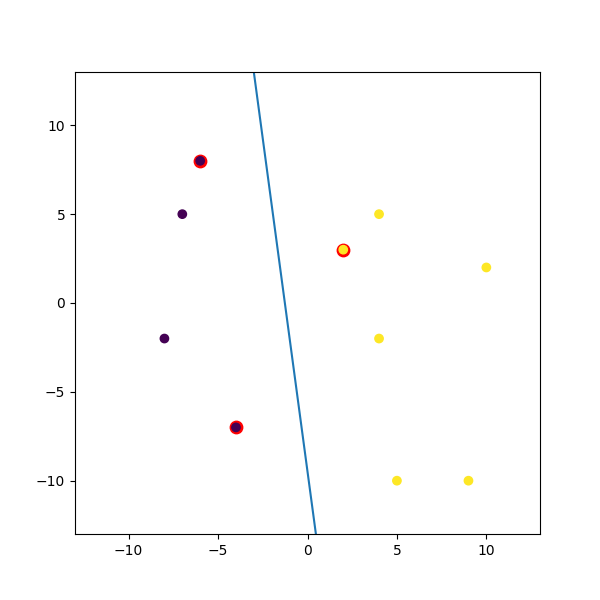
Here is a dummy example I mashed up:
import numpy
import matplotlib.pyplot as plt
from random import seed
from random import randrange
import svmutil as svm
seed(1)
# Creating Data (Dense)
train = list([randrange(-10, 11), randrange(-10, 11)] for i in range(10))
labels = [-1, -1, -1, 1, 1, -1, 1, 1, 1, 1]
options = '-t 0' # linear model
# Training Model
model = svm.svm_train(labels, train, options)
# Line Parameters
w = numpy.matmul(numpy.array(train)[numpy.array(model.get_sv_indices()) - 1].T, model.get_sv_coef())
b = -model.rho.contents.value
if model.get_labels()[1] == -1: # No idea here but it should be done :|
w = -w
b = -b
print(w)
print(b)
# Plotting
plt.figure(figsize=(6, 6))
for i in model.get_sv_indices():
plt.scatter(train[i - 1][0], train[i - 1][1], color='red', s=80)
train = numpy.array(train).T
plt.scatter(train[0], train[1], c=labels)
plt.plot([-5, 5], [-(-5 * w[0] + b) / w[1], -(5 * w[0] + b) / w[1]])
plt.xlim([-13, 13])
plt.ylim([-13, 13])
plt.show()
param = svm_parameter('-s 0 -t 2 -d 3 -c '+str(C)+' -g '+str(G)+' -p '+str(self.epsilon)+' -n '+str(self.nu))
I don't know about the earlier versions but in LibSVM 3.xx the method svm_parameter('options') will takes just one argument.
In my case C, G, p and nu are the dynamic values. You make changes according to your code.
options:
-s svm_type : set type of SVM (default 0)
0 -- C-SVC (multi-class classification)
1 -- nu-SVC (multi-class classification)
2 -- one-class SVM
3 -- epsilon-SVR (regression)
4 -- nu-SVR (regression)
-t kernel_type : set type of kernel function (default 2)
0 -- linear: u'*v
1 -- polynomial: (gamma*u'*v + coef0)^degree
2 -- radial basis function: exp(-gamma*|u-v|^2)
3 -- sigmoid: tanh(gamma*u'*v + coef0)
4 -- precomputed kernel (kernel values in training_set_file)
-d degree : set degree in kernel function (default 3)
-g gamma : set gamma in kernel function (default 1/num_features)
-r coef0 : set coef0 in kernel function (default 0)
-c cost : set the parameter C of C-SVC, epsilon-SVR, and nu-SVR (default 1)
-n nu : set the parameter nu of nu-SVC, one-class SVM, and nu-SVR (default 0.5)
-p epsilon : set the epsilon in loss function of epsilon-SVR (default 0.1)
-m cachesize : set cache memory size in MB (default 100)
-e epsilon : set tolerance of termination criterion (default 0.001)
-h shrinking : whether to use the shrinking heuristics, 0 or 1 (default 1)
-b probability_estimates : whether to train a SVC or SVR model for probability estimates, 0 or 1 (default 0)
-wi weight : set the parameter C of class i to weight*C, for C-SVC (default 1)
-v n: n-fold cross validation mode
-q : quiet mode (no outputs)
Source of documentation: https://www.csie.ntu.edu.tw/~cjlin/libsvm/
© 2022 - 2024 — McMap. All rights reserved.