- Since
seaborn is a high-level API for matplotlib, this seems to mirror functionality in matplotlib
- According to an example in the JointGrid documentation, the parameter is
fc. To use fc, ec should also be used.
- Specifying
fc='none', without specifying ec, will result in blank markers.
fc: facecolor, ec: edgecolor
'None' and 'none' both work, but not None.- Tested in
python 3.11.3, matplotlib 3.7.1, seaborn 0.12.2
import seaborn as sns
# load data
df = sns.load_dataset("penguins", cache=False)
# plot
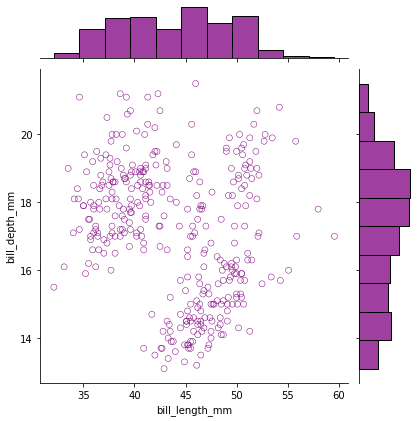
g = sns.jointplot(data=df, x="bill_length_mm", y="bill_depth_mm",
ec="purple", fc="none", color='purple')
![enter image description here]()
- When
hue is used in seaborn v0.12, fc= doesn't seem to work.
- Changing the legend handles comes from this answer.
import seaborn as sns
import matplotlib as mpl
# load data
df = sns.load_dataset("penguins")
# create a palette dict with a known color_palette
species = df.species.unique()
palette = dict(zip(species, sns.color_palette(palette='crest', n_colors=len(species))))
# ec requires a single value or a list of values
ec = df.species.map(palette)
# plot
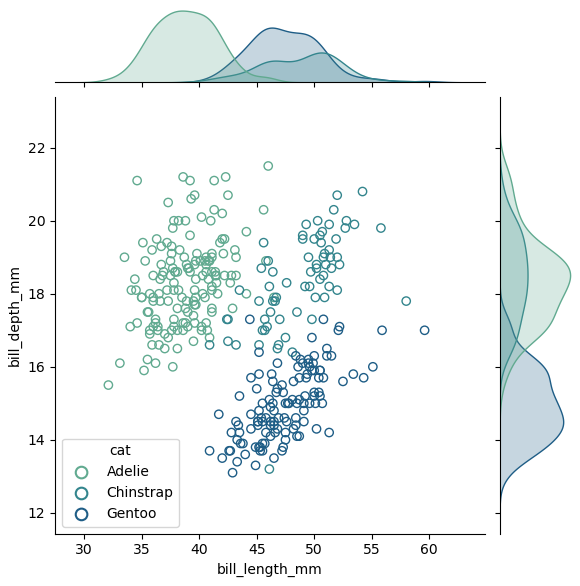
g = sns.jointplot(data=df, x="bill_length_mm", y="bill_depth_mm", ec=ec, hue='species', palette=palette, linewidth=1)
# get the join axes; not the margins
ax_joint = g.ax_joint
# iterate throught axes children
for c in ax_joint.get_children():
# set the facecolor to none
if type(c) == mpl.collections.PathCollection:
c.set_facecolor('none')
# also change the legend
kws = {"s": 70, "facecolor": "none", "linewidth": 1.5}
handles, labels = zip(*[
(plt.scatter([], [], ec=color, **kws), key) for key, color in palette.items()
])
ax_joint.legend(handles, labels, title="cat")
![enter image description here]()
- Using
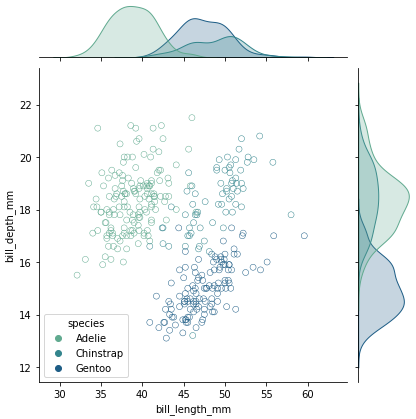
marker="$\circ$" produces this plot.
g = sns.jointplot(data=df, x="bill_length_mm", y="bill_depth_mm", hue='species', palette=palette, marker="$\circ$", s=100)
- In
seaborn v0.12 this doesn't seem to work
- As pointed out in the answer from a11,
ec requires more than a single color if using the hue= parameter. However, it's easier to create palette by zipping the unique values from the column passed to hue, to a known color palette, for anything more than a couple of colors.
palette = dict(zip(df.species.unique(), sns.color_palette('tab10')))
species = df.species.unique() and palette = dict(zip(species, sns.color_palette('crest', n_colors=len(species))))
- If using continuous palettes, specifying
n_colors will generate a palette with better color differentiation.
# plot
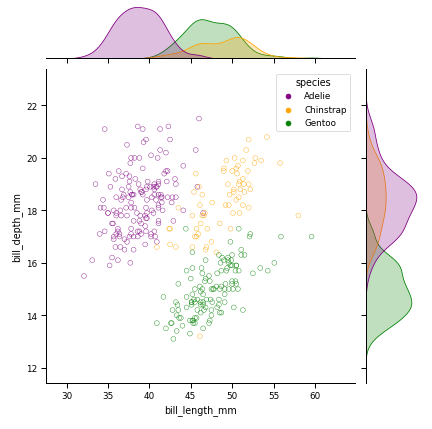
g = sns.jointplot(data=df, x="bill_length_mm", y="bill_depth_mm",
hue='species', ec=ec, fc="none", palette=palette)
![enter image description here]()
Palettes
'Accent', 'Accent_r', 'Blues', 'Blues_r', 'BrBG', 'BrBG_r', 'BuGn', 'BuGn_r', 'BuPu'
'BuPu_r', 'CMRmap', 'CMRmap_r', 'Dark2', 'Dark2_r', 'GnBu', 'GnBu_r', 'Greens', 'Greens_r'
'Greys', 'Greys_r', 'OrRd', 'OrRd_r', 'Oranges', 'Oranges_r', 'PRGn', 'PRGn_r', 'Paired'
'Paired_r', 'Pastel1', 'Pastel1_r', 'Pastel2', 'Pastel2_r', 'PiYG', 'PiYG_r', 'PuBu'
'PuBuGn', 'PuBuGn_r', 'PuBu_r', 'PuOr', 'PuOr_r', 'PuRd', 'PuRd_r', 'Purples', 'Purples_r'
'RdBu', 'RdBu_r', 'RdGy', 'RdGy_r', 'RdPu', 'RdPu_r', 'RdYlBu', 'RdYlBu_r', 'RdYlGn'
'RdYlGn_r', 'Reds', 'Reds_r', 'Set1', 'Set1_r', 'Set2', 'Set2_r', 'Set3', 'Set3_r'
'Spectral', 'Spectral_r', 'Wistia', 'Wistia_r', 'YlGn', 'YlGnBu', 'YlGnBu_r', 'YlGn_r'
'YlOrBr', 'YlOrBr_r', 'YlOrRd', 'YlOrRd_r', 'afmhot', 'afmhot_r', 'autumn', 'autumn_r'
'binary', 'binary_r', 'bone', 'bone_r', 'brg', 'brg_r', 'bwr', 'bwr_r', 'cividis'
'cividis_r', 'cool', 'cool_r', 'coolwarm', 'coolwarm_r', 'copper', 'copper_r', 'crest'
'crest_r', 'cubehelix', 'cubehelix_r', 'flag', 'flag_r', 'flare', 'flare_r', 'gist_earth'
'gist_earth_r', 'gist_gray', 'gist_gray_r', 'gist_heat', 'gist_heat_r', 'gist_ncar'
'gist_ncar_r', 'gist_rainbow', 'gist_rainbow_r', 'gist_stern', 'gist_stern_r', 'gist_yarg'
'gist_yarg_r', 'gnuplot', 'gnuplot2', 'gnuplot2_r', 'gnuplot_r', 'gray', 'gray_r'
'hot', 'hot_r', 'hsv', 'hsv_r', 'icefire', 'icefire_r', 'inferno', 'inferno_r', 'jet'
'jet_r', 'magma', 'magma_r', 'mako', 'mako_r', 'nipy_spectral', 'nipy_spectral_r'
'ocean', 'ocean_r', 'pink', 'pink_r', 'plasma', 'plasma_r', 'prism', 'prism_r', 'rainbow'
'rainbow_r', 'rocket', 'rocket_r', 'seismic', 'seismic_r', 'spring', 'spring_r', 'summer'
'summer_r', 'tab10', 'tab10_r', 'tab20', 'tab20_r', 'tab20b', 'tab20b_r', 'tab20c'
'tab20c_r', 'terrain', 'terrain_r', 'turbo', 'turbo_r', 'twilight', 'twilight_r', 'twilight_shifted'
'twilight_shifted_r', 'viridis', 'viridis_r', 'vlag', 'vlag_r', 'winter', 'winter_r'