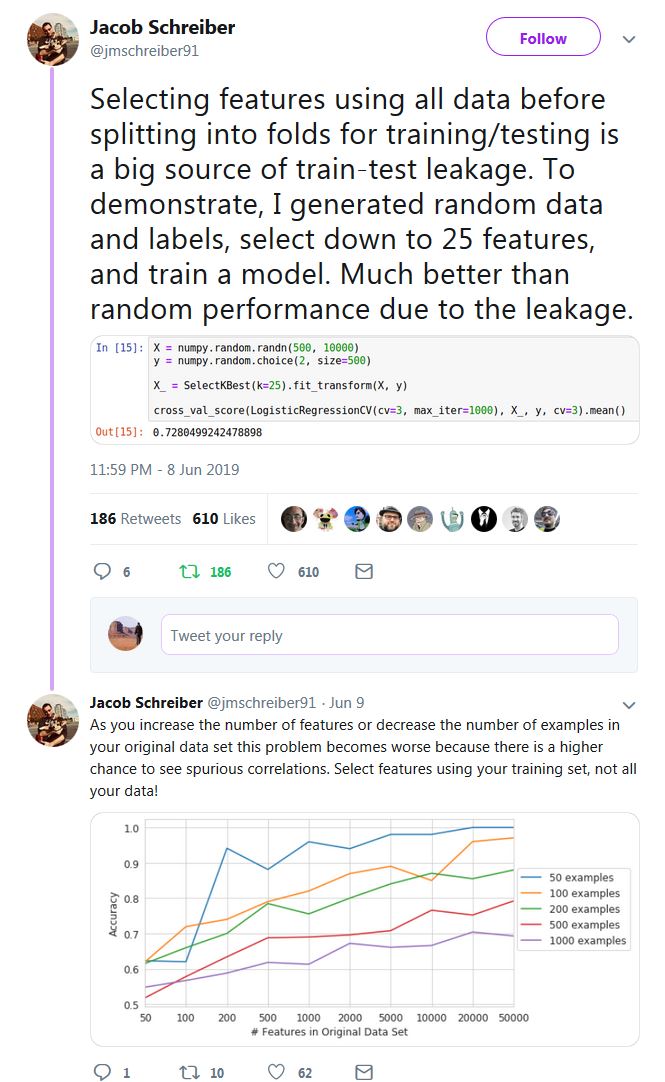
It is not actually difficult to demonstrate why using the whole dataset (i.e. before splitting to train/test) for selecting features can lead you astray. Here is one such demonstration using random dummy data with Python and scikit-learn:
import numpy as np
from sklearn.feature_selection import SelectKBest
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
# random data:
X = np.random.randn(500, 10000)
y = np.random.choice(2, size=500)
Since our data X are random ones (500 samples, 10,000 features) and our labels y are binary, we expect than we should never be able to exceed the baseline accuracy for such a setting, i.e. ~ 0.5, or around 50%. Let's see what happens when we apply the wrong procedure of using the whole dataset for feature selection, before splitting:
selector = SelectKBest(k=25)
# first select features
X_selected = selector.fit_transform(X,y)
# then split
X_selected_train, X_selected_test, y_train, y_test = train_test_split(X_selected, y, test_size=0.25, random_state=42)
# fit a simple logistic regression
lr = LogisticRegression()
lr.fit(X_selected_train,y_train)
# predict on the test set and get the test accuracy:
y_pred = lr.predict(X_selected_test)
accuracy_score(y_test, y_pred)
# 0.76000000000000001
Wow! We get 76% test accuracy on a binary problem where, according to the very basic laws of statistics, we should be getting something very close to 50%! Someone to call the Nobel Prize committee, and fast...
... the truth of course is that we were able to get such a test accuracy simply because we have committed a very basic mistake: we mistakenly think that our test data is unseen, but in fact the test data have already been seen by the model building process during feature selection, in particular here:
X_selected = selector.fit_transform(X,y)
How badly off can we be in reality? Well, again it is not difficult to see: suppose that, after we have finished with our model and we have deployed it (expecting something similar to 76% accuracy in practice with new unseen data), we get some really new data:
X_new = np.random.randn(500, 10000)
where of course there is not any qualitative change, i.e. new trends or anything - these new data are generated by the very same underlying procedure. Suppose also we happen to know the true labels y, generated as above:
y_new = np.random.choice(2, size=500)
How will our model perform here, when faced with these really unseen data? Not difficult to check:
# select the same features in the new data
X_new_selected = selector.transform(X_new)
# predict and get the accuracy:
y_new_pred = lr.predict(X_new_selected)
accuracy_score(y_new, y_new_pred)
# 0.45200000000000001
Well, it's true: we sent our model to the battle, thinking that it was capable of a ~ 76% accuracy, but in reality it performs just as a random guess...
So, let's see now the correct procedure (i.e. split first, and select the features based on the training set only):
# split first
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, random_state=42)
# then select features using the training set only
selector = SelectKBest(k=25)
X_train_selected = selector.fit_transform(X_train,y_train)
# fit again a simple logistic regression
lr.fit(X_train_selected,y_train)
# select the same features on the test set, predict, and get the test accuracy:
X_test_selected = selector.transform(X_test)
y_pred = lr.predict(X_test_selected)
accuracy_score(y_test, y_pred)
# 0.52800000000000002
Where the test accuracy 0f 0.528 is close enough to the theoretically predicted one of 0.5 in such a case (i.e. actually random guessing).
Kudos to Jacob Schreiber for providing the simple idea (check all the thread, it contains other useful examples), although in a slightly different context than the one you ask about here (cross-validation):
![enter image description here]()