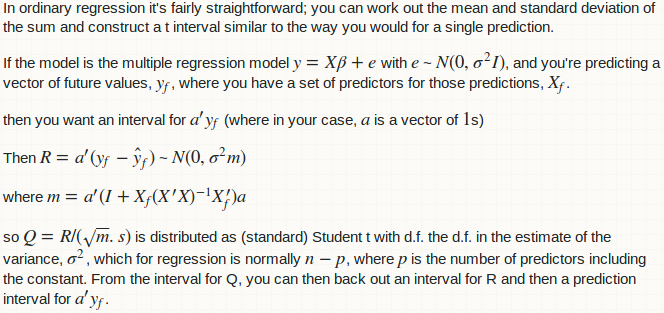
Your question is closely related to a thread I answered 2 years ago: linear model with `lm`: how to get prediction variance of sum of predicted values. It provides an R implementation of Glen_b's answer on Cross Validated. Thanks for quoting that Cross Validated thread; I didn't know it; perhaps I can leave a comment there linking the Stack Overflow thread.
I have polished my original answer, wrapping up line-by-line code cleanly into easy-to-use functions lm_predict and agg_pred. Solving your question is then simplified to applying those functions by group.
Consider the iris example in your question, and the second model fit2 for demonstration.
set.seed(123)
data(iris)
#Split dataset in training and prediction set
smp_size <- floor(0.75 * nrow(iris))
train_ind <- sample(seq_len(nrow(iris)), size = smp_size)
train <- iris[train_ind, ]
pred <- iris[-train_ind, ]
#Fit multiple linear regression model
fit2 <- lm(Petal.Width ~ Petal.Length + Sepal.Width + Sepal.Length, data=train)
We split pred by group Species, then apply lm_predict (with diag = FALSE) on all sub data frames.
oo <- lapply(split(pred, pred$Species), lm_predict, lmObject = fit2, diag = FALSE)
To use agg_pred we need to specify a weight vector, whose length equals to the number of data. We can determine this by consulting the length of fit in each oo[[i]]:
n <- lengths(lapply(oo, "[[", 1))
#setosa versicolor virginica
# 11 13 14
If aggregation operation is sum, we do
w <- lapply(n, rep.int, x = 1)
#List of 3
# $ setosa : num [1:11] 1 1 1 1 1 1 1 1 1 1 ...
# $ versicolor: num [1:13] 1 1 1 1 1 1 1 1 1 1 ...
# $ virginica : num [1:14] 1 1 1 1 1 1 1 1 1 1 ...
SUM <- Map(agg_pred, w, oo)
SUM[[1]] ## result for the first group, for example
#$mean
#[1] 2.499728
#
#$var
#[1] 0.1271554
#
#$CI
# lower upper
#1.792908 3.206549
#
#$PI
# lower upper
#0.999764 3.999693
sapply(SUM, "[[", "CI") ## some nice presentation for CI, for example
# setosa versicolor virginica
#lower 1.792908 16.41526 26.55839
#upper 3.206549 17.63953 28.10812
If aggregation operation is average, we rescale w by n and call agg_pred.
w <- mapply("/", w, n)
#List of 3
# $ setosa : num [1:11] 0.0909 0.0909 0.0909 0.0909 0.0909 ...
# $ versicolor: num [1:13] 0.0769 0.0769 0.0769 0.0769 0.0769 ...
# $ virginica : num [1:14] 0.0714 0.0714 0.0714 0.0714 0.0714 ...
AVE <- Map(agg_pred, w, oo)
AVE[[2]] ## result for the second group, for example
#$mean
#[1] 1.3098
#
#$var
#[1] 0.0005643196
#
#$CI
# lower upper
#1.262712 1.356887
#
#$PI
# lower upper
#1.189562 1.430037
sapply(AVE, "[[", "PI") ## some nice presentation for CI, for example
# setosa versicolor virginica
#lower 0.09088764 1.189562 1.832255
#upper 0.36360845 1.430037 2.072496
This is great! Thank you so much! There is one thing I forgot to mention: in my actual application I need to sum ~300,000 predictions which would create a full variance-covariance matrix which is about ~700GB in size. Do you have any idea if there is a computationally more efficient way to directly get to the sum of the variance-covariance matrix?
Use the fast_agg_pred function provided in the revision of the original Q & A. Let's start it all over.
set.seed(123)
data(iris)
#Split dataset in training and prediction set
smp_size <- floor(0.75 * nrow(iris))
train_ind <- sample(seq_len(nrow(iris)), size = smp_size)
train <- iris[train_ind, ]
pred <- iris[-train_ind, ]
#Fit multiple linear regression model
fit2 <- lm(Petal.Width ~ Petal.Length + Sepal.Width + Sepal.Length, data=train)
## list of new data
newdatlist <- split(pred, pred$Species)
n <- sapply(newdatlist, nrow)
#setosa versicolor virginica
# 11 13 14
If aggregation operation is sum, we do
w <- lapply(n, rep.int, x = 1)
SUM <- mapply(fast_agg_pred, w, newdatlist,
MoreArgs = list(lmObject = fit2, alpha = 0.95),
SIMPLIFY = FALSE)
If aggregation operation is average, we do
w <- mapply("/", w, n)
AVE <- mapply(fast_agg_pred, w, newdatlist,
MoreArgs = list(lmObject = fit2, alpha = 0.95),
SIMPLIFY = FALSE)
Note that we can't use Map in this case as we need to provide more arguments to fast_agg_pred. Use mapply in this situation, with MoreArgs and SIMPLIFY.