What is the fastest way to check if a value exists in a very large list (with millions of values) and what its index is?
7 in a
Clearest and fastest way to do it.
You can also consider using a set, but constructing that set from your list may take more time than faster membership testing will save. The only way to be certain is to benchmark well. (this also depends on what operations you require)
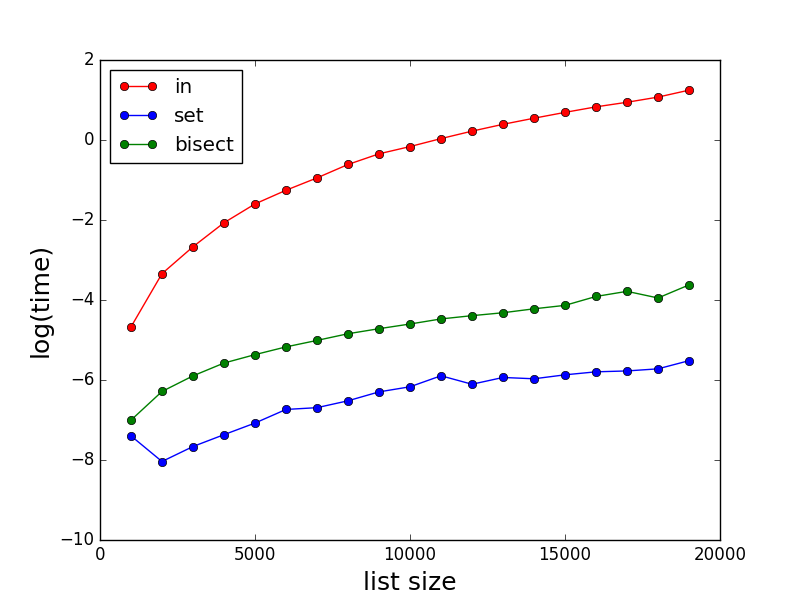
As stated by others, in can be very slow for large lists. Here are some comparisons of the performances for in, set and bisect. Note the time (in second) is in log scale.
Code for testing:
import random
import bisect
import matplotlib.pyplot as plt
import math
import time
def method_in(a, b, c):
start_time = time.time()
for i, x in enumerate(a):
if x in b:
c[i] = 1
return time.time() - start_time
def method_set_in(a, b, c):
start_time = time.time()
s = set(b)
for i, x in enumerate(a):
if x in s:
c[i] = 1
return time.time() - start_time
def method_bisect(a, b, c):
start_time = time.time()
b.sort()
for i, x in enumerate(a):
index = bisect.bisect_left(b, x)
if index < len(a):
if x == b[index]:
c[i] = 1
return time.time() - start_time
def profile():
time_method_in = []
time_method_set_in = []
time_method_bisect = []
# adjust range down if runtime is too long or up if there are too many zero entries in any of the time_method lists
Nls = [x for x in range(10000, 30000, 1000)]
for N in Nls:
a = [x for x in range(0, N)]
random.shuffle(a)
b = [x for x in range(0, N)]
random.shuffle(b)
c = [0 for x in range(0, N)]
time_method_in.append(method_in(a, b, c))
time_method_set_in.append(method_set_in(a, b, c))
time_method_bisect.append(method_bisect(a, b, c))
plt.plot(Nls, time_method_in, marker='o', color='r', linestyle='-', label='in')
plt.plot(Nls, time_method_set_in, marker='o', color='b', linestyle='-', label='set')
plt.plot(Nls, time_method_bisect, marker='o', color='g', linestyle='-', label='bisect')
plt.xlabel('list size', fontsize=18)
plt.ylabel('log(time)', fontsize=18)
plt.legend(loc='upper left')
plt.yscale('log')
plt.show()
profile()
import random / import bisect / import matplotlib.pyplot as plt and then call: profile() –
Barrington range() object. When using var in [integer list], see if a range() object can model the same sequence. Very close in performance to a set, but more concise. –
Unrobe x in list check. –
Sp You could put your items into a set. Set lookups are very efficient.
Try:
s = set(a)
if 7 in s:
# do stuff
edit In a comment you say that you'd like to get the index of the element. Unfortunately, sets have no notion of element position. An alternative is to pre-sort your list and then use binary search every time you need to find an element.
The original question was:
What is the fastest way to know if a value exists in a list (a list with millions of values in it) and what its index is?
Thus there are two things to find:
- is an item in the list, and
- what is the index (if in the list).
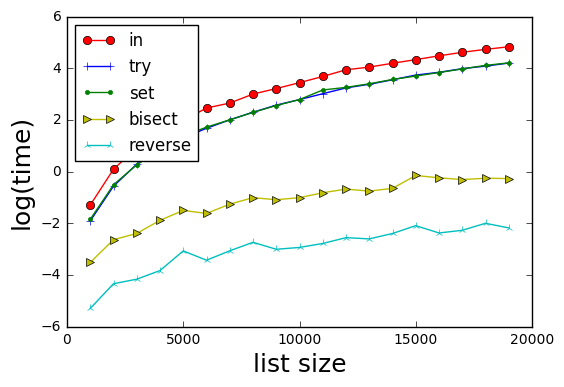
Towards this, I modified @xslittlegrass code to compute indexes in all cases, and added an additional method.
Results
Methods are:
- in--basically if x in b: return b.index(x)
- try--try/catch on b.index(x) (skips having to check if x in b)
- set--basically if x in set(b): return b.index(x)
- bisect--sort b with its index, binary search for x in sorted(b). Note mod from @xslittlegrass who returns the index in the sorted b, rather than the original b)
- reverse--form a reverse lookup dictionary d for b; then d[x] provides the index of x.
Results show that method 5 is the fastest.
Interestingly the try and the set methods are equivalent in time.
Test Code
import random
import bisect
import matplotlib.pyplot as plt
import math
import timeit
import itertools
def wrapper(func, *args, **kwargs):
" Use to produced 0 argument function for call it"
# Reference https://www.pythoncentral.io/time-a-python-function/
def wrapped():
return func(*args, **kwargs)
return wrapped
def method_in(a,b,c):
for i,x in enumerate(a):
if x in b:
c[i] = b.index(x)
else:
c[i] = -1
return c
def method_try(a,b,c):
for i, x in enumerate(a):
try:
c[i] = b.index(x)
except ValueError:
c[i] = -1
def method_set_in(a,b,c):
s = set(b)
for i,x in enumerate(a):
if x in s:
c[i] = b.index(x)
else:
c[i] = -1
return c
def method_bisect(a,b,c):
" Finds indexes using bisection "
# Create a sorted b with its index
bsorted = sorted([(x, i) for i, x in enumerate(b)], key = lambda t: t[0])
for i,x in enumerate(a):
index = bisect.bisect_left(bsorted,(x, ))
c[i] = -1
if index < len(a):
if x == bsorted[index][0]:
c[i] = bsorted[index][1] # index in the b array
return c
def method_reverse_lookup(a, b, c):
reverse_lookup = {x:i for i, x in enumerate(b)}
for i, x in enumerate(a):
c[i] = reverse_lookup.get(x, -1)
return c
def profile():
Nls = [x for x in range(1000,20000,1000)]
number_iterations = 10
methods = [method_in, method_try, method_set_in, method_bisect, method_reverse_lookup]
time_methods = [[] for _ in range(len(methods))]
for N in Nls:
a = [x for x in range(0,N)]
random.shuffle(a)
b = [x for x in range(0,N)]
random.shuffle(b)
c = [0 for x in range(0,N)]
for i, func in enumerate(methods):
wrapped = wrapper(func, a, b, c)
time_methods[i].append(math.log(timeit.timeit(wrapped, number=number_iterations)))
markers = itertools.cycle(('o', '+', '.', '>', '2'))
colors = itertools.cycle(('r', 'b', 'g', 'y', 'c'))
labels = itertools.cycle(('in', 'try', 'set', 'bisect', 'reverse'))
for i in range(len(time_methods)):
plt.plot(Nls,time_methods[i],marker = next(markers),color=next(colors),linestyle='-',label=next(labels))
plt.xlabel('list size', fontsize=18)
plt.ylabel('log(time)', fontsize=18)
plt.legend(loc = 'upper left')
plt.show()
profile()
def check_availability(element, collection: iter):
return element in collection
Usage
check_availability('a', [1,2,3,4,'a','b','c'])
I believe this is the fastest way to know if a chosen value is in an array.
o='--skip'; o in ("--skip-ias"); # returns True ! –
Janise in operator works the same way to test substring membership. The confusing part here is probably that ("hello") is not a single-value tuple, while ("hello",) is -- the comma makes the difference. o in ("--skip-ias",) is False as expected. –
Chacon a = [4,2,3,1,5,6]
index = dict((y,x) for x,y in enumerate(a))
try:
a_index = index[7]
except KeyError:
print "Not found"
else:
print "found"
This will only be a good idea if a doesn't change and thus we can do the dict() part once and then use it repeatedly. If a does change, please provide more detail on what you are doing.
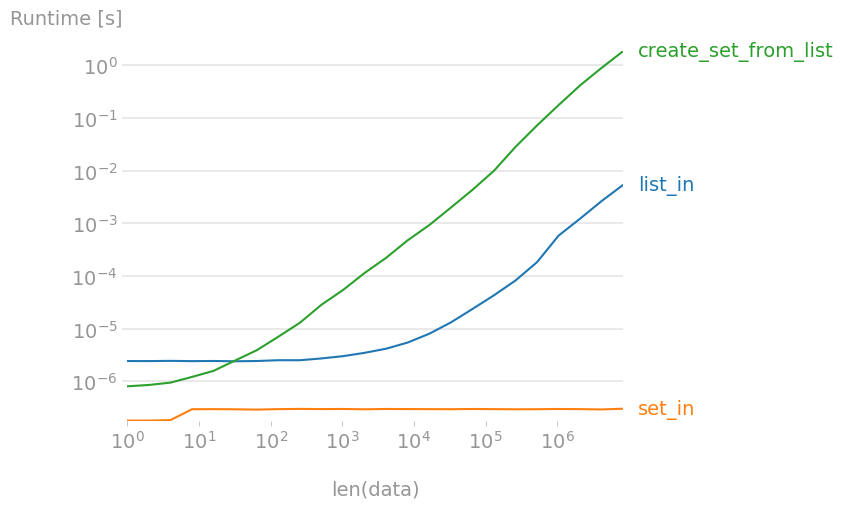
If you only want to check the existence of one element in a list,
7 in list_data
is the fastest solution. Note though that
7 in set_data
is a near-free operation, independently of the size of the set! Creating a set from a large list is 300 to 400 times slower than in, so if you need to check for many elements, creating a set first is faster.
Plot created with perfplot:
import perfplot
import numpy as np
def setup(n):
data = np.arange(n)
np.random.shuffle(data)
return data, set(data)
def list_in(data):
return 7 in data[0]
def create_set_from_list(data):
return set(data[0])
def set_in(data):
return 7 in data[1]
b = perfplot.bench(
setup=setup,
kernels=[list_in, set_in, create_set_from_list],
n_range=[2 ** k for k in range(24)],
xlabel="len(data)",
equality_check=None,
)
b.save("out.png")
b.show()
set IMO AFAIK. –
Sur Be aware that the in operator tests not only equality (==) but also identity (is), the in logic for lists is roughly equivalent to the following (it's actually written in C and not Python though, at least in CPython):
for element in s: if element is target: # fast check for identity implies equality return True if element == target: # slower check for actual equality return True return False
In most circumstances this detail is irrelevant, but in some circumstances it might leave a Python novice surprised, for example, numpy.NAN has the unusual property of being not being equal to itself:
>>> import numpy
>>> numpy.NAN == numpy.NAN
False
>>> numpy.NAN is numpy.NAN
True
>>> numpy.NAN in [numpy.NAN]
True
To distinguish between these unusual cases you could use any() like:
>>> lst = [numpy.NAN, 1 , 2]
>>> any(element == numpy.NAN for element in lst)
False
>>> any(element is numpy.NAN for element in lst)
True
Note the in logic for lists with any() would be:
any(element is target or element == target for element in lst)
However, I should emphasize that this is an edge case, and for the vast majority of cases the in operator is highly optimised and exactly what you want of course (either with a list or with a set).
It sounds like your application might gain advantage from the use of a Bloom Filter data structure.
In short, a bloom filter look-up can tell you very quickly if a value is DEFINITELY NOT present in a set. Otherwise, you can do a slower look-up to get the index of a value that POSSIBLY MIGHT BE in the list. So if your application tends to get the "not found" result much more often then the "found" result, you might see a speed up by adding a Bloom Filter.
For details, Wikipedia provides a good overview of how Bloom Filters work, and a web search for "python bloom filter library" will provide at least a couple useful implementations.
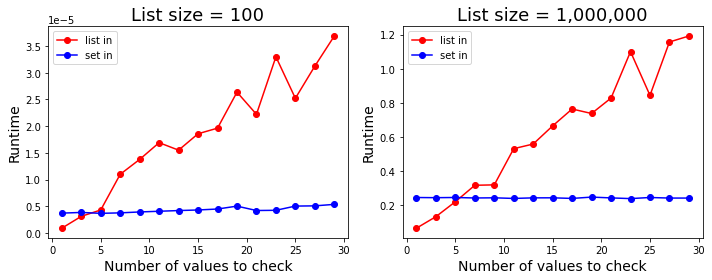
Check if values exist in a list
xslittlegrass's answer shows that when checking if multiple values exist in a list, converting the list into a set first and using the in operator on the set is much faster than using the in operator on lists. On the other hand, Nico's answer shows that when checking if a single value exists in a list, converting the list into a set first is not worth it, as converting to a set itself is costly to begin with. Together, these answers imply that there is some number of values where converting to a set and checking if those values exist in the set becomes faster than checking if they exist in the list.
It turns out, that number is very small. The figure below shows the runtime difference between in on sets and in on lists for different numbers of values to check. As it shows, on average, if you need to check whether 5 (or more) values exist in a list, it's faster to convert that list into a set first and check on the set instead.1
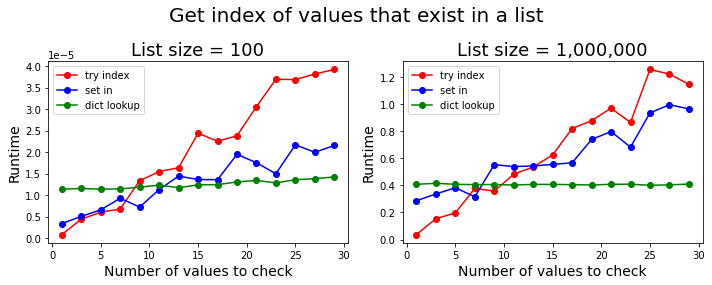
Get their indices if values exist in a list
On the other hand, if you want to check if values exist in a list and return the indices of the values that do, then regardless of the length of the list, for small number of values, directly searching for it using list.index() in a try-except block is the fastest way to do it. In particular, if you want to find the index of a single value, this is the fastest option. However, on average, if there are more than 10 values to search for, then constructing an index lookup dictionary (as in DarrylG's answer) is the fastest option.2
1 Code used to produce the first figure.
from random import sample
from timeit import repeat
from functools import partial
import matplotlib.pyplot as plt
from matplotlib.ticker import MultipleLocator
def list_in(a, b):
return [x in b for x in a]
def set_in(a, b):
s = set(b)
return [x in s for x in a]
def profile(methods):
sizes = range(1, 31, 2)
colors = ['r', 'b', 'g']
Ns = [100, 1000000]
fig, axs = plt.subplots(1, len(Ns), figsize=(10, 4), facecolor='white')
for N, ax in zip(Ns, axs):
b = sample(range(N), k=N)
times = {f.__name__: [] for f in methods}
for size in sizes:
a = sample(range(len(b)*3//2), k=size)
for label, f in zip(times, methods):
func = partial(f, a, b)
times[label].append(min(repeat(func, number=10))/10)
for (k, ts), c in zip(times.items(), colors):
ax.plot(sizes, ts, f'{c}o-', label=k)
ax.set_title(f'List size = {N:,d}', fontsize=18)
ax.set_xlabel('Number of values to check', fontsize=14)
ax.set_ylabel('Runtime', fontsize=14)
ax.xaxis.set_major_locator(MultipleLocator(5))
ax.legend()
return fig
methods = [list_in, set_in]
fig = profile(methods)
fig.tight_layout();
2 Code used to produce the second figure.
def try_index(a, b):
c = []
for x in a:
try:
c.append(b.index(x))
except ValueError:
c.append(-1)
return c
def set_in(a, b):
s = set(b)
return [b.index(x) if x in s else -1 for x in a]
def dict_lookup(a, b):
# for faster lookups, convert dict to a function beforehand
reverse_lookup = {x:i for i, x in enumerate(b)}.get
return [reverse_lookup(x, -1) for x in a]
methods = [try_index, set_in, dict_lookup]
fig = profile(methods)
fig.suptitle('Get index of values that exist in a list', fontsize=20)
fig.tight_layout();
This is not the code, but the algorithm for very fast searching.
If your list and the value you are looking for are all numbers, this is pretty straightforward. If strings: look at the bottom:
- -Let "n" be the length of your list
- -Optional step: if you need the index of the element: add a second column to the list with current index of elements (0 to n-1) - see later
- Order your list or a copy of it (.sort())
- Loop through:
- Compare your number to the n/2th element of the list
- If larger, loop again between indexes n/2-n
- If smaller, loop again between indexes 0-n/2
- If the same: you found it
- Compare your number to the n/2th element of the list
- Keep narrowing the list until you have found it or only have 2 numbers (below and above the one you are looking for)
- This will find any element in at most 19 steps for a list of 1.000.000 (log(2)n to be precise)
If you also need the original position of your number, look for it in the second, index column.
If your list is not made of numbers, the method still works and will be fastest, but you may need to define a function which can compare/order strings.
Of course, this needs the investment of the sorted() method, but if you keep reusing the same list for checking, it may be worth it.
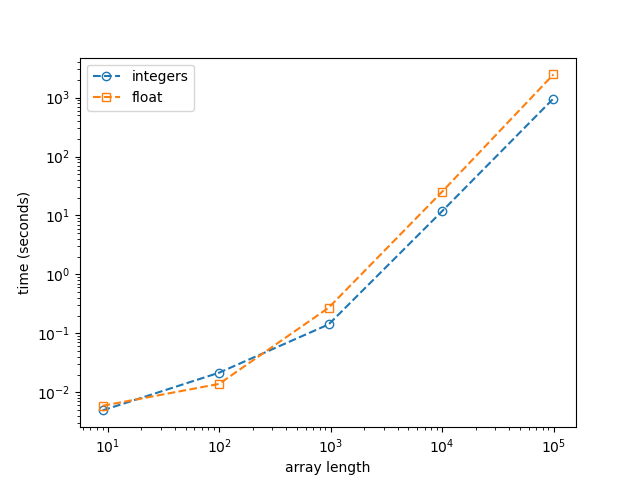
Edge case for spatial data
There are probably faster algorithms for handling spatial data (e.g. refactoring to use a k-d tree), but the special case of checking if a vector is in an array is useful:
- If you have spatial data (i.e. cartesian coordinates)
- If you have integer masks (i.e. array filtering)
In this case, I was interested in knowing if an (undirected) edge defined by two points was in a collection of (undirected) edges, such that
(pair in unique_pairs) | (pair[::-1] in unique_pairs) for pair in pairs
where pair constitutes two vectors of arbitrary length (i.e. shape (2,N)).
If the distance between these vectors is meaningful, then the test can be expressed by a floating point inequality like
test_result = Norm(v1 - v2) < Tol
and "Value exists in List" is simply any(test_result).
Example code and dummy test set generators for integer pairs and R3 vector pairs are below.
# 3rd party
import numpy as np
import numpy.linalg as LA
import matplotlib.pyplot as plt
# optional
try:
from tqdm import tqdm
except ModuleNotFoundError:
def tqdm(X, *args, **kwargs):
return X
print('tqdm not found. tqdm is a handy progress bar module.')
def get_float_r3_pairs(size):
""" generate dummy vector pairs in R3 (i.e. case of spatial data) """
coordinates = np.random.random(size=(size, 3))
pairs = []
for b in coordinates:
for a in coordinates:
pairs.append((a,b))
pairs = np.asarray(pairs)
return pairs
def get_int_pairs(size):
""" generate dummy integer pairs (i.e. case of array masking) """
coordinates = np.random.randint(0, size, size)
pairs = []
for b in coordinates:
for a in coordinates:
pairs.append((a,b))
pairs = np.asarray(pairs)
return pairs
def float_tol_pair_in_pairs(pair:np.ndarray, pairs:np.ndarray) -> np.ndarray:
"""
True if abs(a0 - b0) <= tol & abs(a1 - b1) <= tol for (ai1, aj2), (bi1, bj2)
in [(a01, a02), ... (aik, ajl)]
NB this is expected to be called in iteration so no sanitization is performed.
Parameters
----------
pair : np.ndarray
pair of vectors with shape (2, M)
pairs : np.ndarray
collection of vector pairs with shape (N, 2, M)
Returns
-------
np.ndarray
(pair in pairs) | (pair[::-1] in pairs).
"""
m1 = np.sum( abs(LA.norm(pairs - pair, axis=2)) <= (1e-03, 1e-03), axis=1 ) == 2
m2 = np.sum( abs(LA.norm(pairs - pair[::-1], axis=2)) <= (1e-03, 1e-03), axis=1 ) == 2
return m1 | m2
def get_unique_pairs(pairs:np.ndarray) -> np.ndarray:
"""
apply float_tol_pair_in_pairs for pair in pairs
Parameters
----------
pairs : np.ndarray
collection of vector pairs with shape (N, 2, M)
Returns
-------
np.ndarray
pair if not ((pair in rv) | (pair[::-1] in rv)) for pair in pairs
"""
pairs = np.asarray(pairs).reshape((len(pairs), 2, -1))
rv = [pairs[0]]
for pair in tqdm(pairs[1:], desc='finding unique pairs...'):
if not any(float_tol_pair_in_pairs(pair, rv)):
rv.append(pair)
return np.array(rv)
© 2022 - 2024 — McMap. All rights reserved.

bisectmodule – Triglyceride