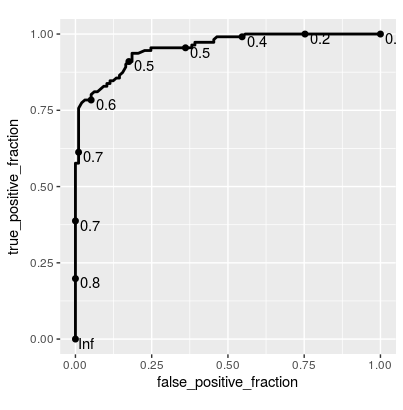
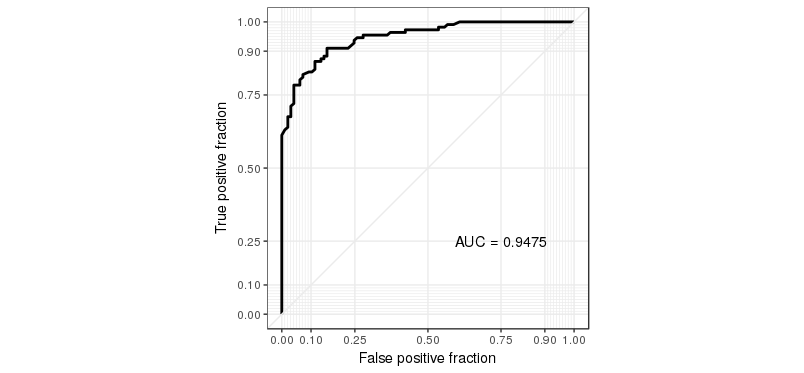
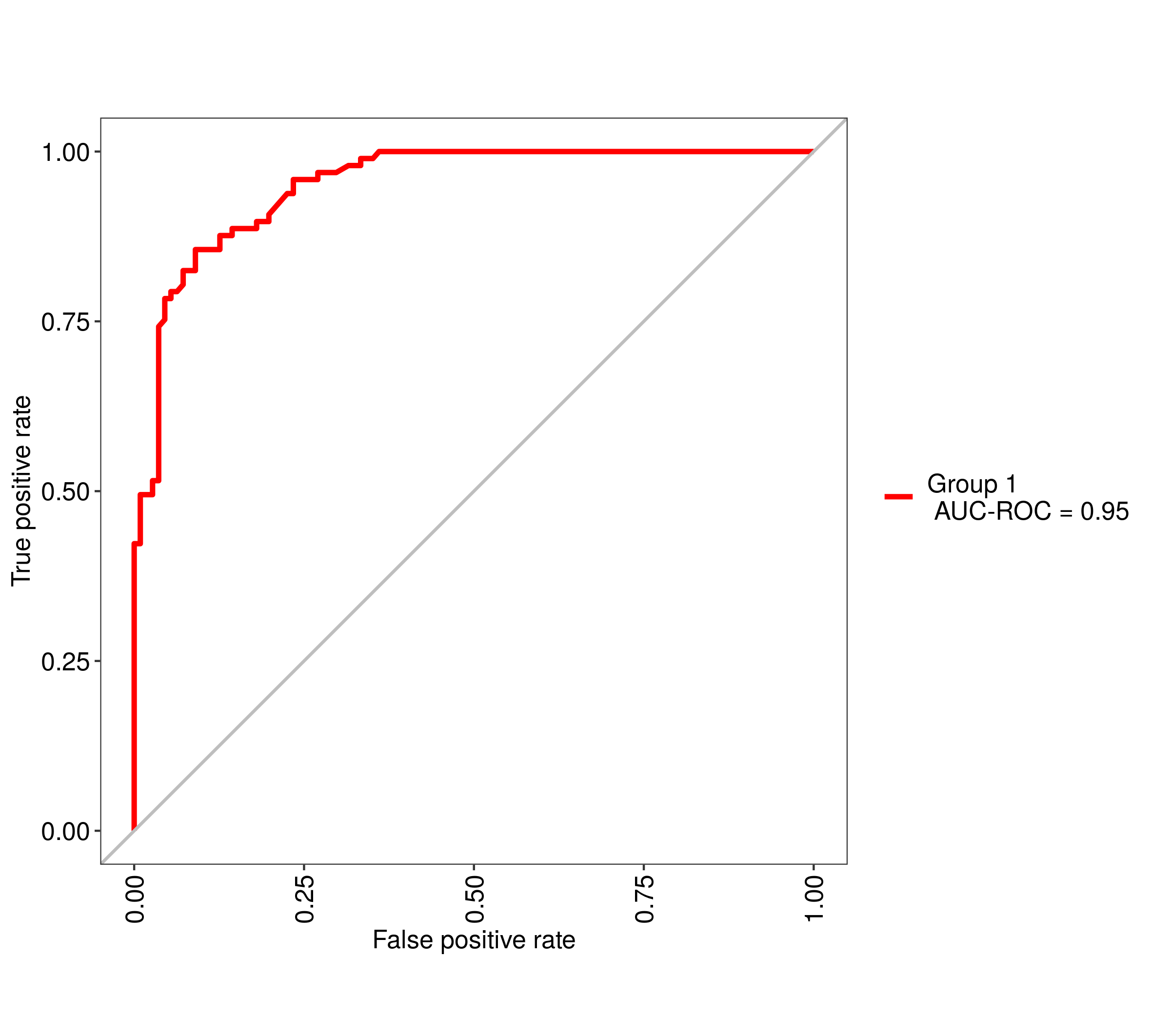
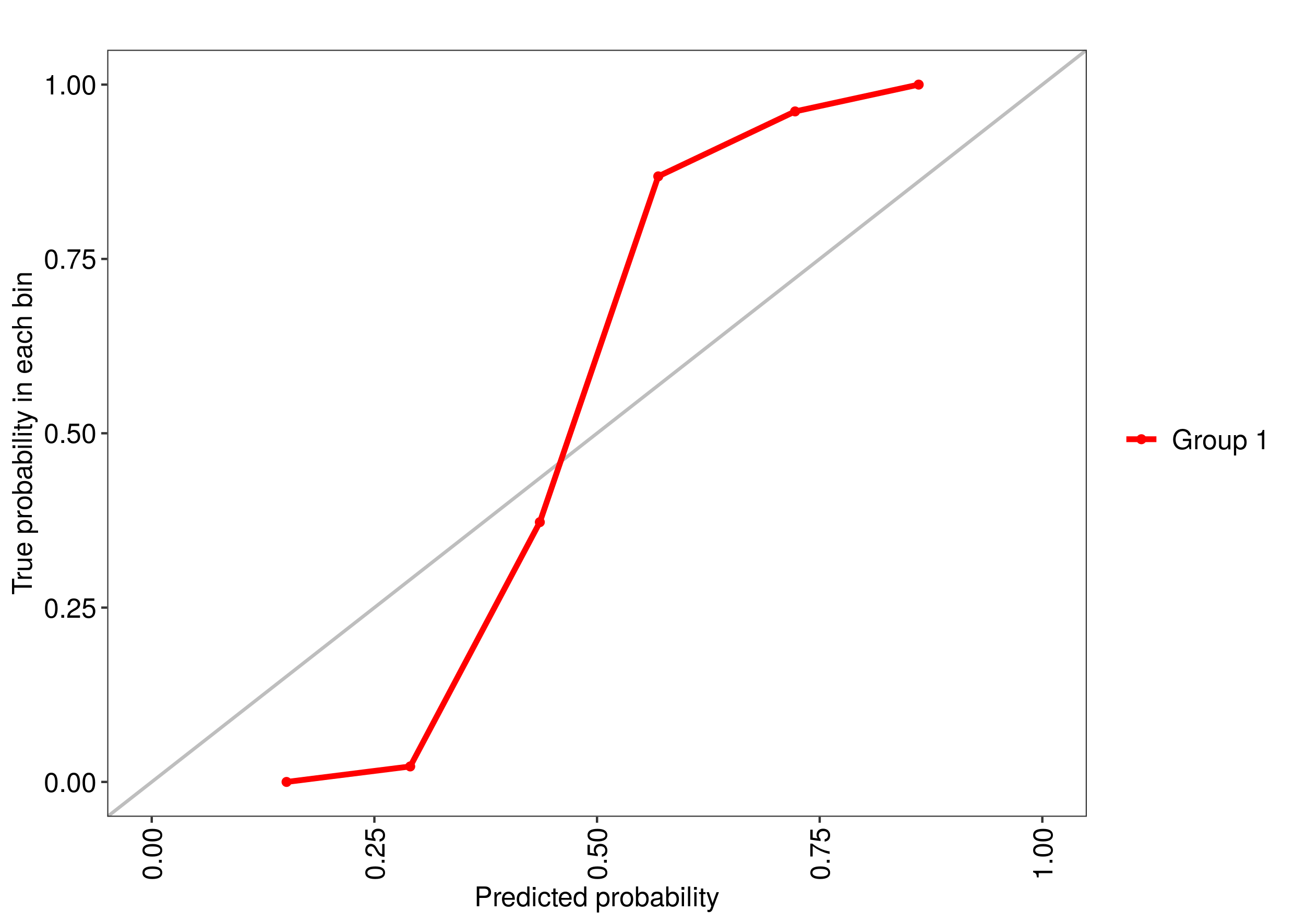
Using the R package caret, how can I generate a ROC curve based on the cross-validation results of the train() function?
Say, I do the following:
data(Sonar)
ctrl <- trainControl(method="cv",
summaryFunction=twoClassSummary,
classProbs=T)
rfFit <- train(Class ~ ., data=Sonar,
method="rf", preProc=c("center", "scale"),
trControl=ctrl)
The training function goes over a range of mtry parameter and calculates the ROC AUC. I would like to see the associated ROC curve -- how do I do that?
Note: if the method used for sampling is LOOCV, then rfFit will contain a non-null data frame in the rfFit$pred slot, which seems to be exactly what I need. However, I need that for the "cv" method (k-fold validation) rather than LOO.
Also: no, roc function that used to be included in former versions of caret is not an answer -- this is a low level function, you can't use it if you don't have the prediction probabilities for each cross-validated sample.