So, to try to put all these reasonable answers together:
(In the following, I used V for the number of vertices in the generated graph, and E for the number of edges, and we assume that E ≤ V(V-1)/2.)
Personally, I think the most useful answer is in a comment, by Flavius, who points at the code at http://condor.depaul.edu/rjohnson/source/graph_ge.c. That code is really simple, and it's conveniently described by a comment, which I reproduce:
To generate a directed acyclic graph, we first
generate a random permutation dag[0],...,dag[v-1].
(v = number of vertices.)
This random permutation serves as a topological
sort of the graph. We then generate random edges of the
form (dag[i],dag[j]) with i < j.
In fact, what the code does is generate the request number of edges by repeatedly doing the following:
- generate two numbers in the range [0, V);
- reject them if they're equal;
- swap them if the first is larger;
- reject them if it has generated them before.
The problem with this solution is that as E gets closes to the maximum number of edges V(V-1)/2, then the algorithm becomes slower and slower, because it has to reject more and more edges. A better solution would be to make a vector of all V(V-1)/2 possible edges; randomly shuffle it; and select the first (requested edges) edges in the shuffled list.
The reservoir sampling algorithm lets us do this in space O(E), since we can deduce the endpoints of the kth edge from the value of k. Consequently, we don't actually have to create the source vector. However, it still requires O(V2) time.
Alternatively, one can do a Fisher-Yates shuffle (or Knuth shuffle, if you prefer), stopping after E iterations. In the version of the FY shuffle presented in Wikipedia, this will produce the trailing entries, but the algorithm works just as well backwards:
// At the end of this snippet, a consists of a random sample of the
// integers in the half-open range [0, V(V-1)/2). (They still need to be
// converted to pairs of endpoints).
vector<int> a;
int N = V * (V - 1) / 2;
for (int i = 0; i < N; ++i) a.push_back(i);
for (int i = 0; i < E; ++i) {
int j = i + rand(N - i);
swap(a[i], a[j]);
a.resize(E);
This requires only O(E) time but it requires O(N2) space. In fact, this can be improved to O(E) space with some trickery, but an SO code snippet is too small to contain the result, so I'll provide a simpler one in O(E) space and O(E log E) time. I assume that there is a class DAG with at least:
class DAG {
// Construct an empty DAG with v vertices
explicit DAG(int v);
// Add the directed edge i->j, where 0 <= i, j < v
void add(int i, int j);
};
Now here goes:
// Return a randomly-constructed DAG with V vertices and and E edges.
// It's required that 0 < E < V(V-1)/2.
template<typename PRNG>
DAG RandomDAG(int V, int E, PRNG& prng) {
using dist = std::uniform_int_distribution<int>;
// Make a random sample of size E
std::vector<int> sample;
sample.reserve(E);
int N = V * (V - 1) / 2;
dist d(0, N - E); // uniform_int_distribution is closed range
// Random vector of integers in [0, N-E]
for (int i = 0; i < E; ++i) sample.push_back(dist(prng));
// Sort them, and make them unique
std::sort(sample.begin(), sample.end());
for (int i = 1; i < E; ++i) sample[i] += i;
// Now it's a unique sorted list of integers in [0, N-E+E-1]
// Randomly shuffle the endpoints, so the topological sort
// is different, too.
std::vector<int> endpoints;
endpoints.reserve(V);
for (i = 0; i < V; ++i) endpoints.push_back(i);
std::shuffle(endpoints.begin(), endpoints.end(), prng);
// Finally, create the dag
DAG rv;
for (auto& v : sample) {
int tail = int(0.5 + sqrt((v + 1) * 2));
int head = v - tail * (tail - 1) / 2;
rv.add(head, tail);
}
return rv;
}


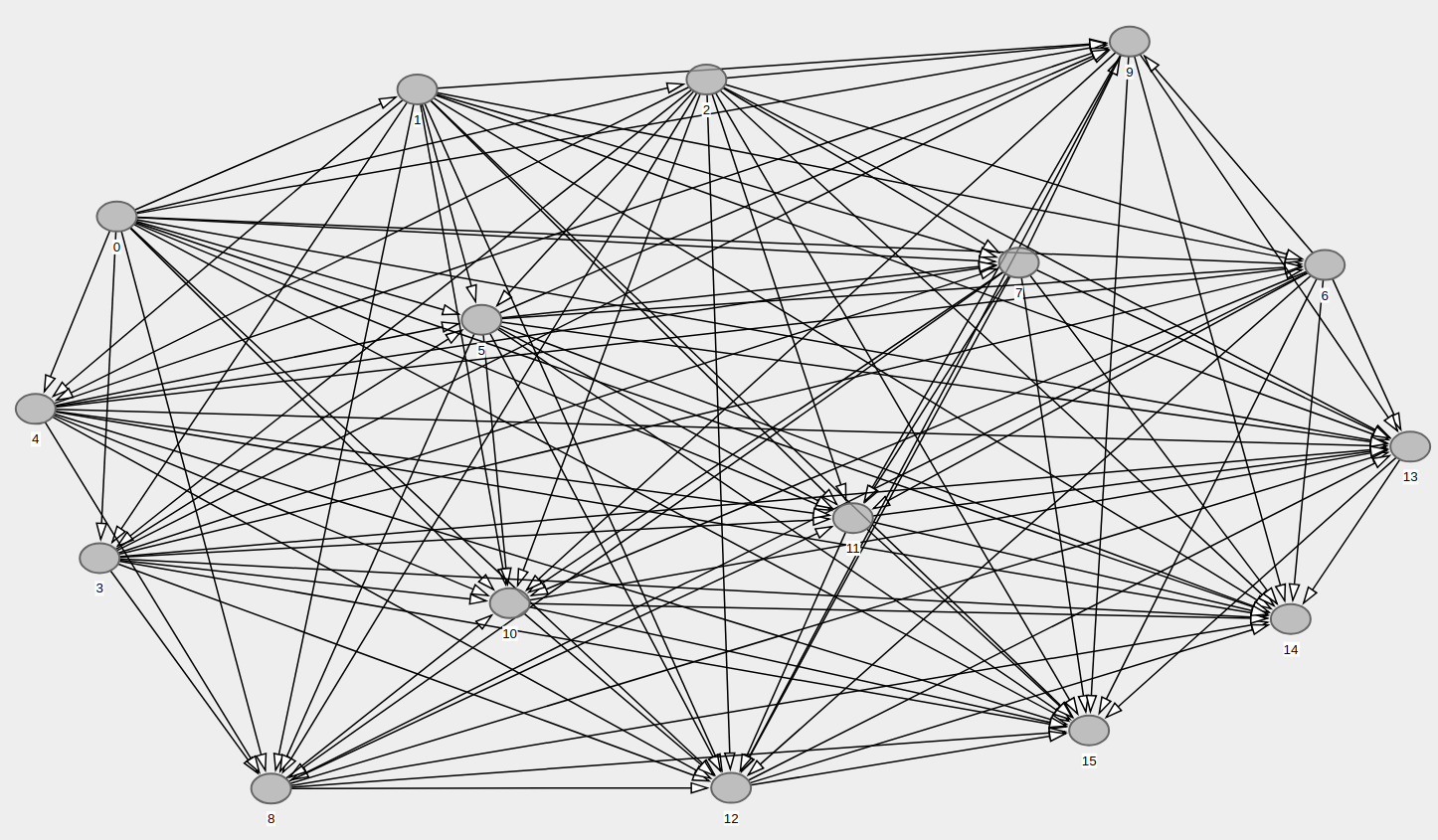
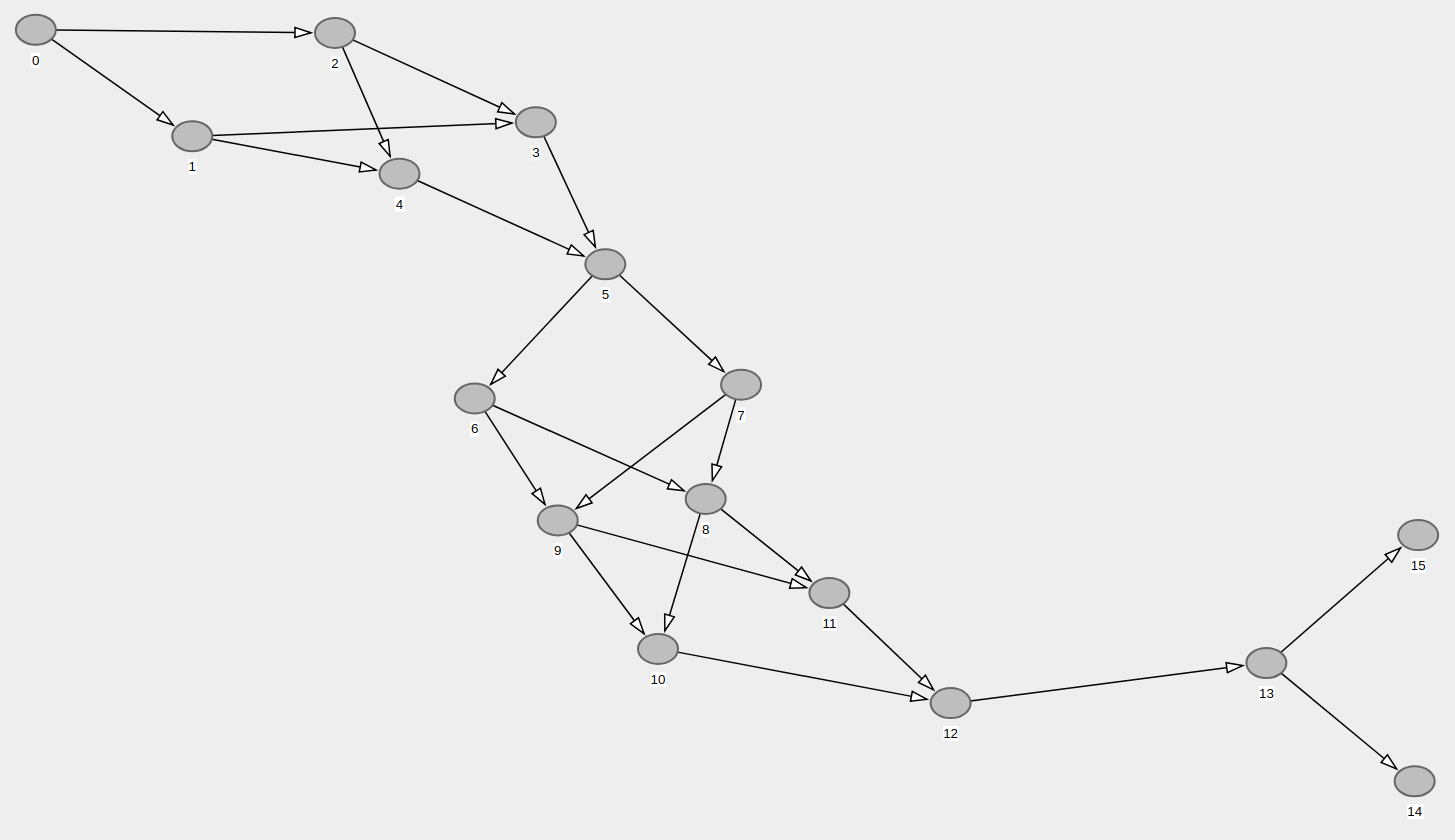
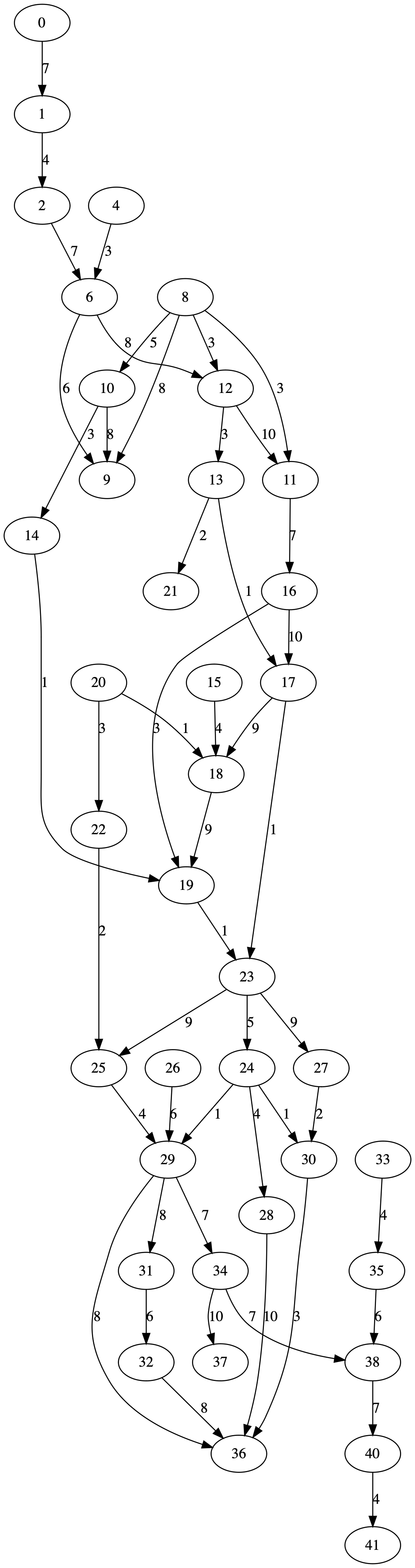

directed_acyclic_graphhere: condor.depaul.edu/rjohnson/source/graph_ge.c – Dornick