I’m trying to fit and plot a Weibull model to a survival data. The data has just one covariate, cohort, which runs from 2006 to 2010. So, any ideas on what to add to the two lines of code that follows to plot the survival curve of the cohort of 2010?
library(survival)
s <- Surv(subSetCdm$dur,subSetCdm$event)
sWei <- survreg(s ~ cohort,dist='weibull',data=subSetCdm)
Accomplishing the same with the Cox PH model is rather straightforward, with the following lines. The problem is that survfit() doesn’t accept objects of type survreg.
sCox <- coxph(s ~ cohort,data=subSetCdm)
cohort <- factor(c(2010),levels=2006:2010)
sfCox <- survfit(sCox,newdata=data.frame(cohort))
plot(sfCox,col='green')
Using the data lung (from the survival package), here is what I'm trying to accomplish.
#create a Surv object
s <- with(lung,Surv(time,status))
#plot kaplan-meier estimate, per sex
fKM <- survfit(s ~ sex,data=lung)
plot(fKM)
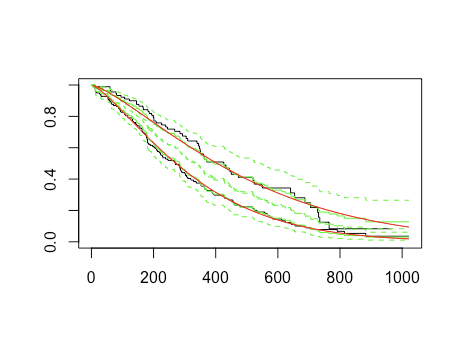
#plot Cox PH survival curves, per sex
sCox <- coxph(s ~ as.factor(sex),data=lung)
lines(survfit(sCox,newdata=data.frame(sex=1)),col='green')
lines(survfit(sCox,newdata=data.frame(sex=2)),col='green')
#plot weibull survival curves, per sex, DOES NOT RUN
sWei <- survreg(s ~ as.factor(sex),dist='weibull',data=lung)
lines(survfit(sWei,newdata=data.frame(sex=1)),col='red')
lines(survfit(sWei,newdata=data.frame(sex=2)),col='red')



?predict.survreg. – Meyers