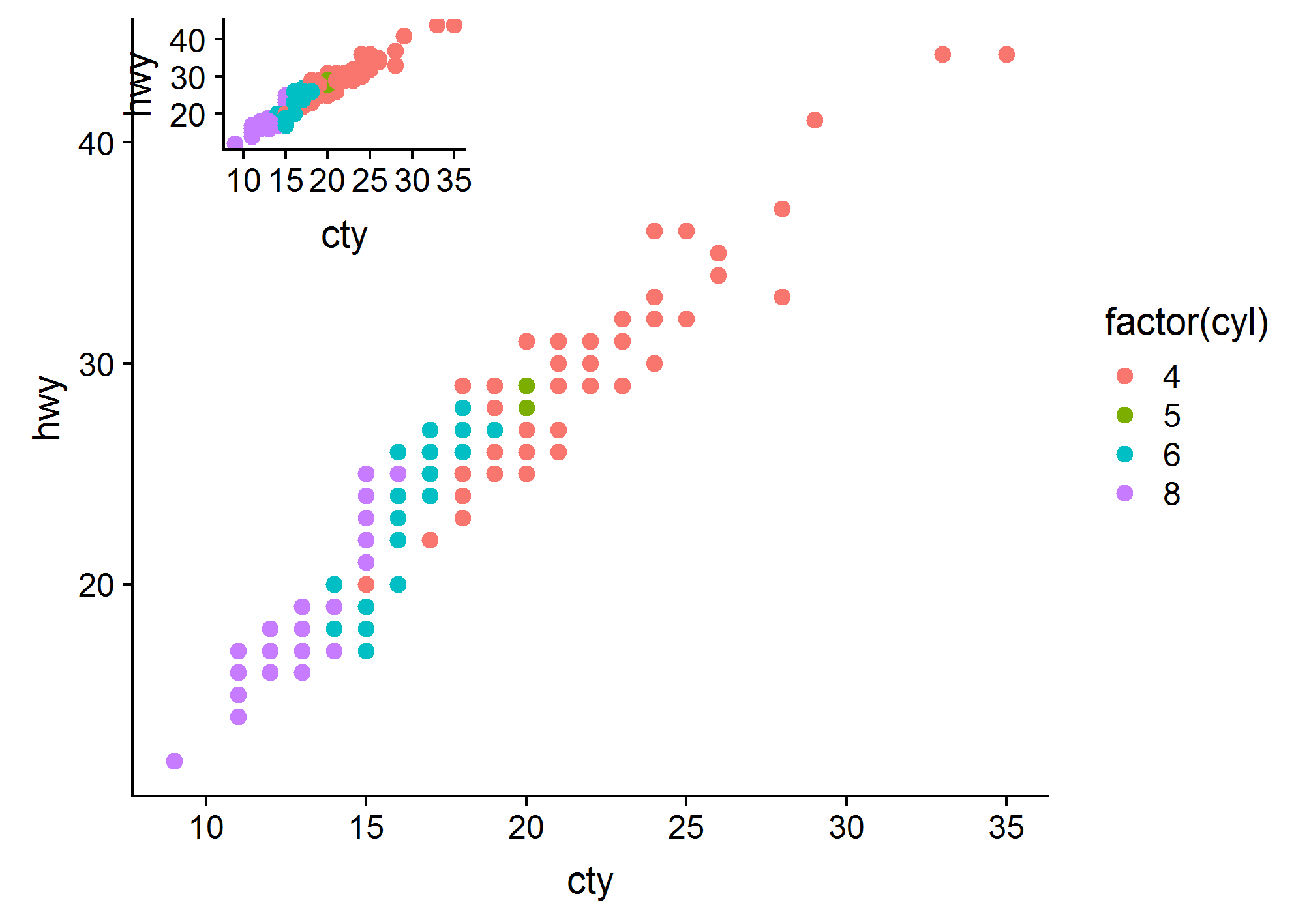
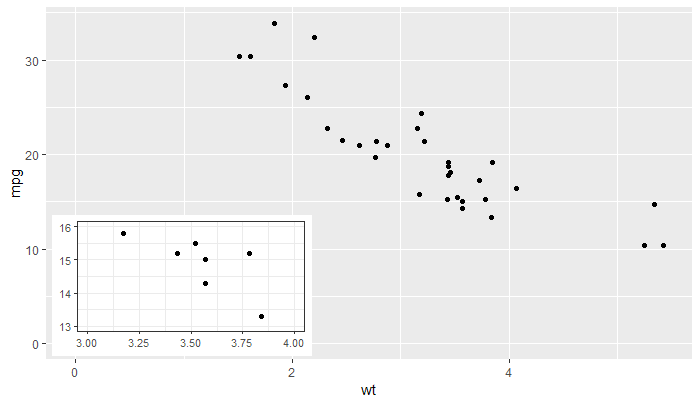
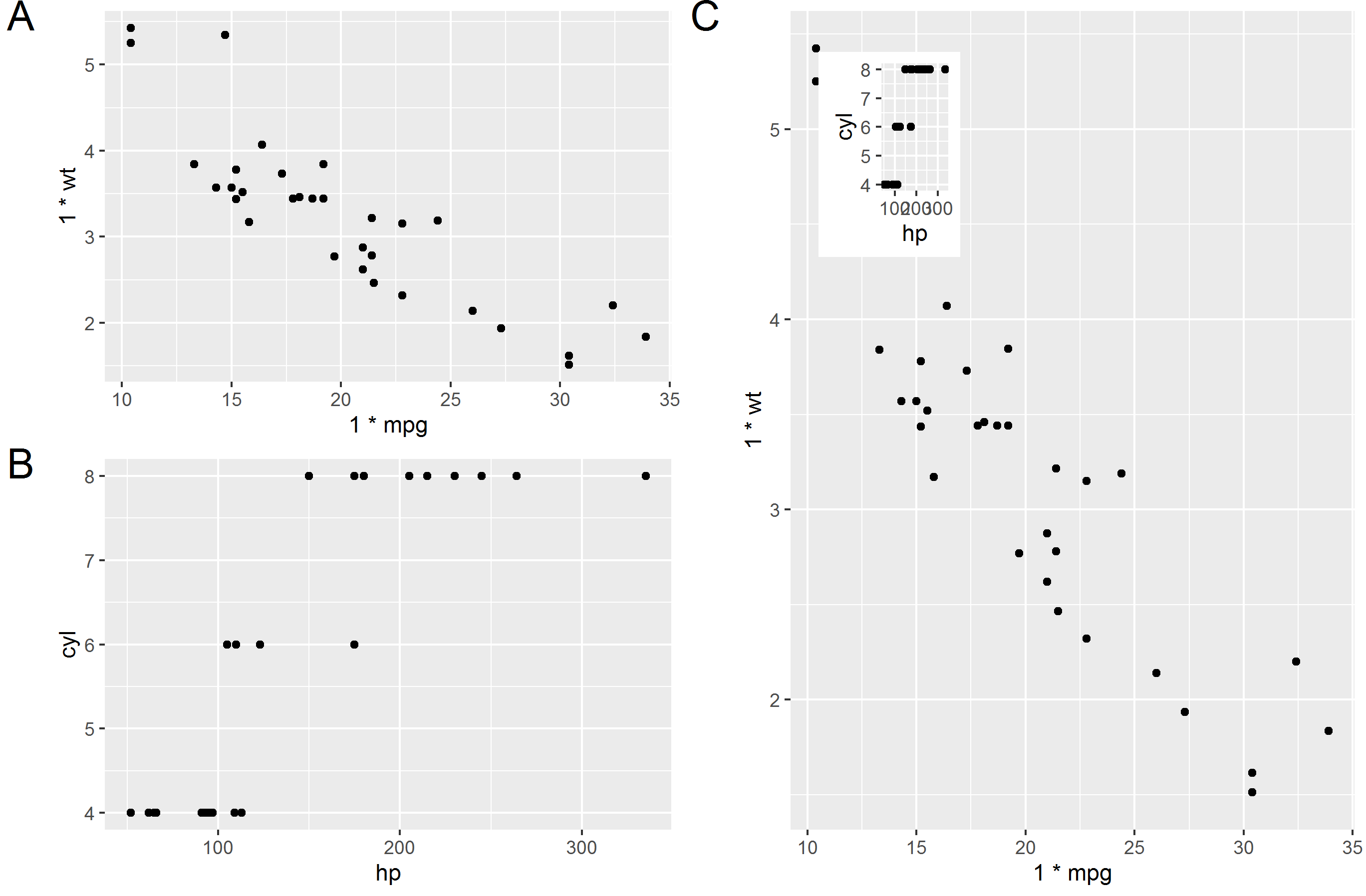
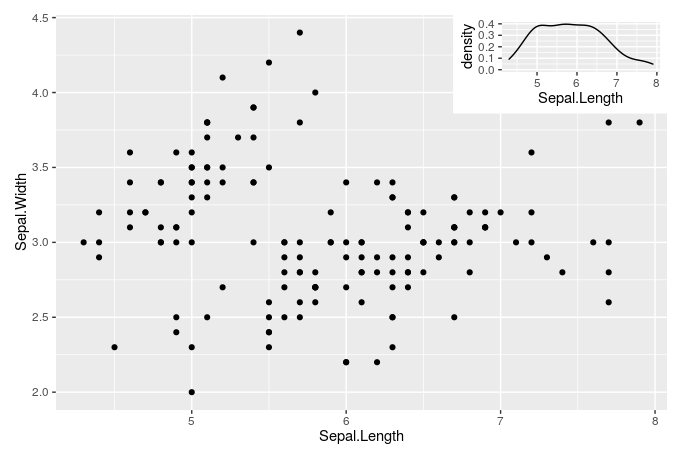
'ggplot2' >= 3.0.0 makes possible new approaches for adding insets, as now tibble objects containing lists as member columns can be passed as data. The objects in the list column can be even whole ggplots... The latest version of my package 'ggpmisc' provides geom_plot(), geom_table() and geom_grob(), and also versions that use npc units instead of native data units for locating the insets. These geoms can add multiple insets per call and obey faceting, which annotation_custom() does not. I copy the example from the help page, which adds an inset with a zoom-in detail of the main plot as an inset.
library(tibble)
library(ggpmisc)
p <-
ggplot(data = mtcars, mapping = aes(wt, mpg)) +
geom_point()
df <- tibble(x = 0.01, y = 0.01,
plot = list(p +
coord_cartesian(xlim = c(3, 4),
ylim = c(13, 16)) +
labs(x = NULL, y = NULL) +
theme_bw(10)))
p +
expand_limits(x = 0, y = 0) +
geom_plot_npc(data = df, aes(npcx = x, npcy = y, label = plot))
![ggplot with ggplot as inset]()
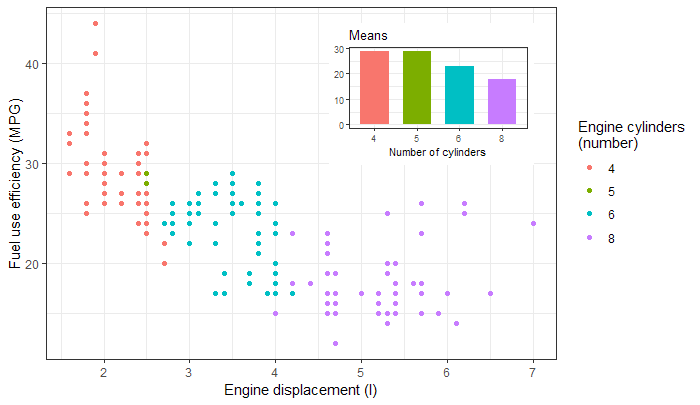
Or a barplot as inset, taken from the package vignette.
library(tibble)
library(ggpmisc)
p <- ggplot(mpg, aes(factor(cyl), hwy, fill = factor(cyl))) +
stat_summary(geom = "col", fun.y = mean, width = 2/3) +
labs(x = "Number of cylinders", y = NULL, title = "Means") +
scale_fill_discrete(guide = FALSE)
data.tb <- tibble(x = 7, y = 44,
plot = list(p +
theme_bw(8)))
ggplot(mpg, aes(displ, hwy, colour = factor(cyl))) +
geom_plot(data = data.tb, aes(x, y, label = plot)) +
geom_point() +
labs(x = "Engine displacement (l)", y = "Fuel use efficiency (MPG)",
colour = "Engine cylinders\n(number)") +
theme_bw()
![enter image description here]()
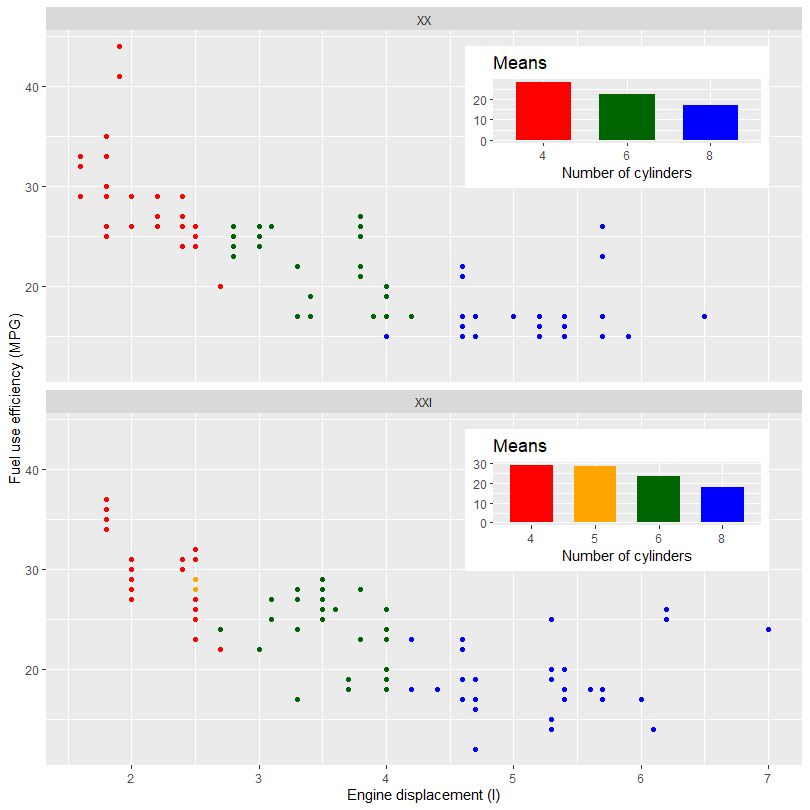
The next example shows how to add different inset plots to different panels in a faceted plot. The next example uses the same example data after splitting it according to the century. This particular data set once split adds the problem of one missing level in one of the inset plots. As these plots are built on their own we need to use manual scales to make sure the colors and fill are consistent across the plots. With other data sets this may not be needed.
library(tibble)
library(ggpmisc)
my.mpg <- mpg
my.mpg$century <- factor(ifelse(my.mpg$year < 2000, "XX", "XXI"))
my.mpg$cyl.f <- factor(my.mpg$cyl)
my_scale_fill <- scale_fill_manual(guide = FALSE,
values = c("red", "orange", "darkgreen", "blue"),
breaks = levels(my.mpg$cyl.f))
p1 <- ggplot(subset(my.mpg, century == "XX"),
aes(factor(cyl), hwy, fill = cyl.f)) +
stat_summary(geom = "col", fun = mean, width = 2/3) +
labs(x = "Number of cylinders", y = NULL, title = "Means") +
my_scale_fill
p2 <- ggplot(subset(my.mpg, century == "XXI"),
aes(factor(cyl), hwy, fill = cyl.f)) +
stat_summary(geom = "col", fun = mean, width = 2/3) +
labs(x = "Number of cylinders", y = NULL, title = "Means") +
my_scale_fill
data.tb <- tibble(x = c(7, 7),
y = c(44, 44),
century = factor(c("XX", "XXI")),
plot = list(p1, p2))
ggplot() +
geom_plot(data = data.tb, aes(x, y, label = plot)) +
geom_point(data = my.mpg, aes(displ, hwy, colour = cyl.f)) +
labs(x = "Engine displacement (l)", y = "Fuel use efficiency (MPG)",
colour = "Engine cylinders\n(number)") +
scale_colour_manual(guide = FALSE,
values = c("red", "orange", "darkgreen", "blue"),
breaks = levels(my.mpg$cyl.f)) +
facet_wrap(~century, ncol = 1)
![enter image description here]()