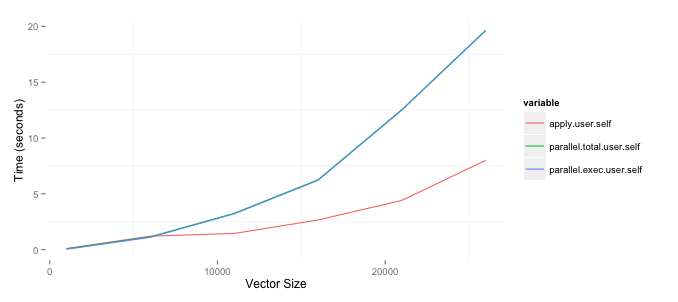
I am trying to determine when to use the parallel package to speed up the time necessary to run some analysis. One of the things I need to do is create matrices comparing variables in two data frames with differing number of rows. I asked a question as to an efficient way of doing on StackOverflow and wrote about tests on my blog. Since I am comfortable with the best approach I wanted to speed up the process by running it in parallel. The results below are based upon a 2ghz i7 Mac with 8gb of RAM. I am surprised that the parallel package, the parSapply funciton in particular, is worse than just using the apply function. The code to replicate this is below. Note that I am currently only using one of the two columns I create but eventually want to use both.
(source: bryer.org)
require(parallel)
require(ggplot2)
require(reshape2)
set.seed(2112)
results <- list()
sizes <- seq(1000, 30000, by=5000)
pb <- txtProgressBar(min=0, max=length(sizes), style=3)
for(cnt in 1:length(sizes)) {
i <- sizes[cnt]
df1 <- data.frame(row.names=1:i,
var1=sample(c(TRUE,FALSE), i, replace=TRUE),
var2=sample(1:10, i, replace=TRUE) )
df2 <- data.frame(row.names=(i + 1):(i + i),
var1=sample(c(TRUE,FALSE), i, replace=TRUE),
var2=sample(1:10, i, replace=TRUE))
tm1 <- system.time({
df6 <- sapply(df2$var1, FUN=function(x) { x == df1$var1 })
dimnames(df6) <- list(row.names(df1), row.names(df2))
})
rm(df6)
tm2 <- system.time({
cl <- makeCluster(getOption('cl.cores', detectCores()))
tm3 <- system.time({
df7 <- parSapply(cl, df1$var1, FUN=function(x, df2) { x == df2$var1 }, df2=df2)
dimnames(df7) <- list(row.names(df1), row.names(df2))
})
stopCluster(cl)
})
rm(df7)
results[[cnt]] <- c(apply=tm1, parallel.total=tm2, parallel.exec=tm3)
setTxtProgressBar(pb, cnt)
}
toplot <- as.data.frame(results)[,c('apply.user.self','parallel.total.user.self',
'parallel.exec.user.self')]
toplot$size <- sizes
toplot <- melt(toplot, id='size')
ggplot(toplot, aes(x=size, y=value, colour=variable)) + geom_line() +
xlab('Vector Size') + ylab('Time (seconds)')

