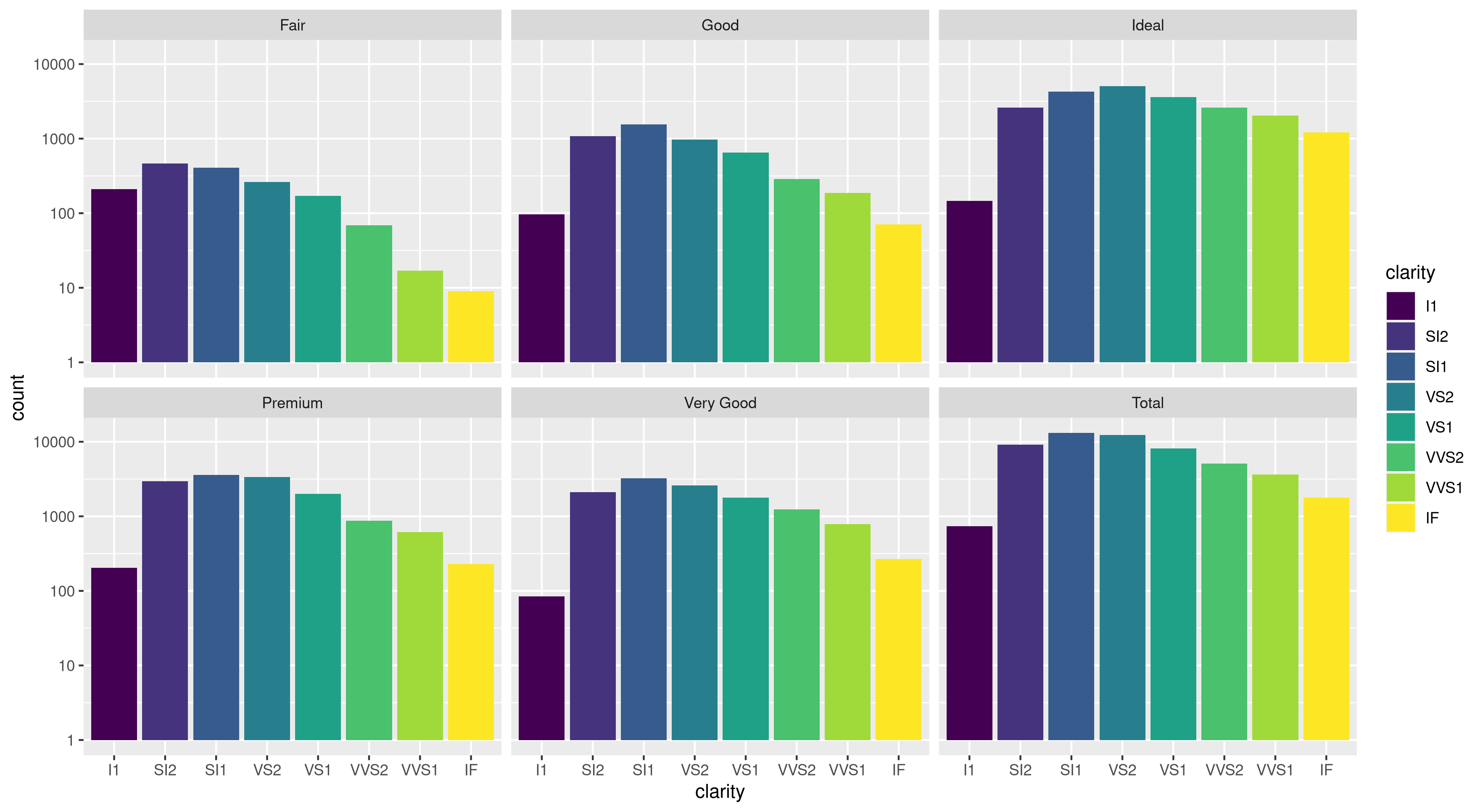
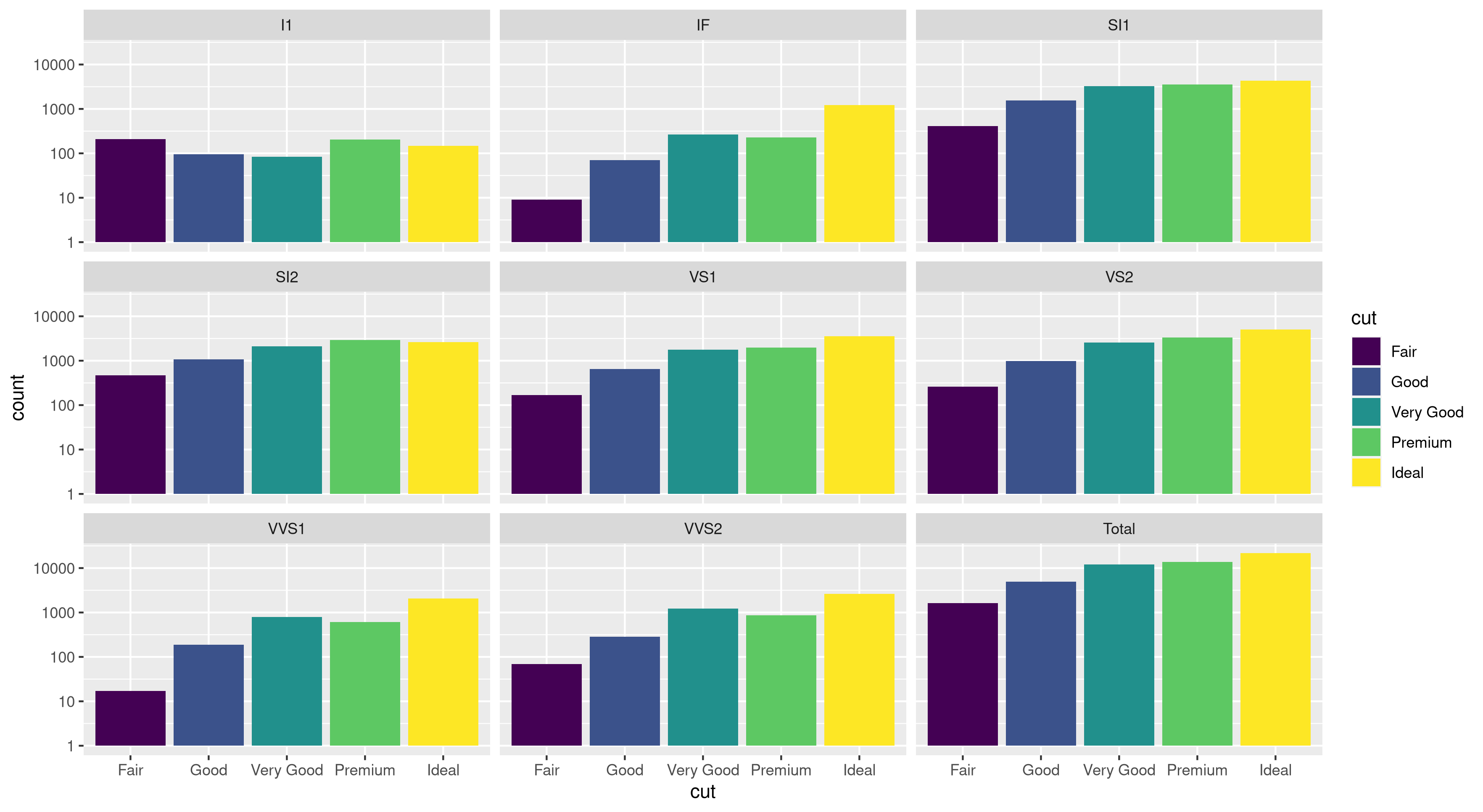
I've run into an interesting problem with scaling using ggplot. I have a dataset that I can graph just fine using the default linear scale but when I use scale_y_log10() the numbers go way off. Here is some example code and two pictures. Note that the max value in the linear scale is ~700 while the log scaling results in a value of 10^8. I show you that the entire dataset is only ~8000 entries long so something is not right.
I imagine the problem has something to do with the structure of my dataset and the binning as I cannot replicate this error on a common dataset like 'diamonds.' However I am not sure the best way to troubleshoot.
thanks, zach cp
Edit: bdamarest can reproduce the scale problem on the diamond dataset like this:
example_1 = ggplot(diamonds, aes(x=clarity, fill=cut)) +
geom_bar() + scale_y_log10(); print(example_1)
#data.melt is the name of my dataset
> ggplot(data.melt, aes(name, fill= Library)) + geom_bar()
> ggplot(data.melt, aes(name, fill= Library)) + geom_bar() + scale_y_log10()
> length(data.melt$name)
[1] 8003


here is some example data... and I think I see the problem. The original melted dataset may have been ~10^8 rows long. Maybe the row numbers are being used for the stats?
> head(data.melt)
Library name group
221938 AB Arthrofactin glycopeptide
235087 AB Putisolvin cyclic peptide
235090 AB Putisolvin cyclic peptide
222125 AB Arthrofactin glycopeptide
311468 AB Triostin cyclic depsipeptide
92249 AB CDA lipopeptide
test2 <- data.frame(
Library = rep("AB", 6L),
name = c(
"Arthrofactin", "Putisolvin", "Putisolvin", "Arthrofactin",
"Triostin", "CDA"
),
group = c(
"glycopeptide", "cyclic peptide", "cyclic peptide", "glycopeptide",
"cyclic depsipeptide", "lipopeptide"
),
row.names = c(221938L, 235087L, 235090L, 222125L, 311468L, 92249L)
)
UPDATE:
Row numbers are not the issue. Here is the same data graphed using the same aes x-axis and fill color and the scaling is entirely correct:
> ggplot(data.melt, aes(name, fill= name)) + geom_bar()
> ggplot(data.melt, aes(name, fill= name)) + geom_bar() + scale_y_log10()
> length(data.melt$name)
[1] 8003