I apologize for the long answer in advance :P (the problem is not that simple).
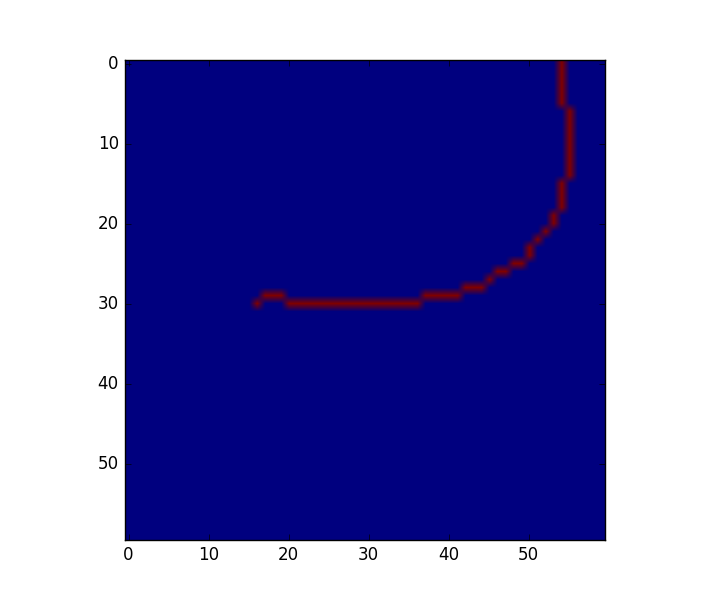
Lets start by rewording the problem. Finding a line that connects all the points, can be reformulated as a shortest path problem in a graph, where (1) the graph nodes are the points in the space, (2) each node is connected to its 2 nearest neighbors, and (3) the shortest path passes through each of the nodes only once. That last constrain is a very important (and quite hard one to optimize). Essentially, the problem is to find a permutation of length N, where the permutation refers to the order of each of the nodes (N is the total number of nodes) in the path.
Finding all the possible permutations and evaluating their cost is too expensive (there are N! permutations if I'm not wrong, which is too big for problems). Bellow I propose an approach that finds the N best permutations (the optimal permutation for each of the N points) and then find the permutation (from those N) that minimizes the error/cost.
1. Create a random problem with unordered points
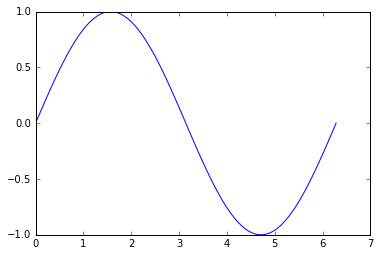
Now, lets start to create a sample problem:
import matplotlib.pyplot as plt
import numpy as np
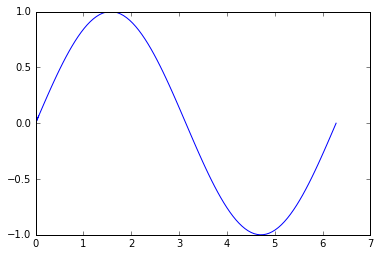
x = np.linspace(0, 2 * np.pi, 100)
y = np.sin(x)
plt.plot(x, y)
plt.show()
![enter image description here]()
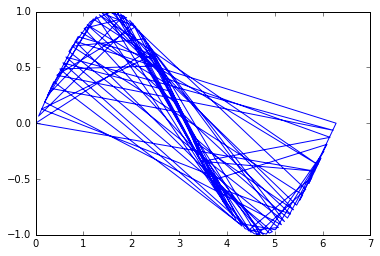
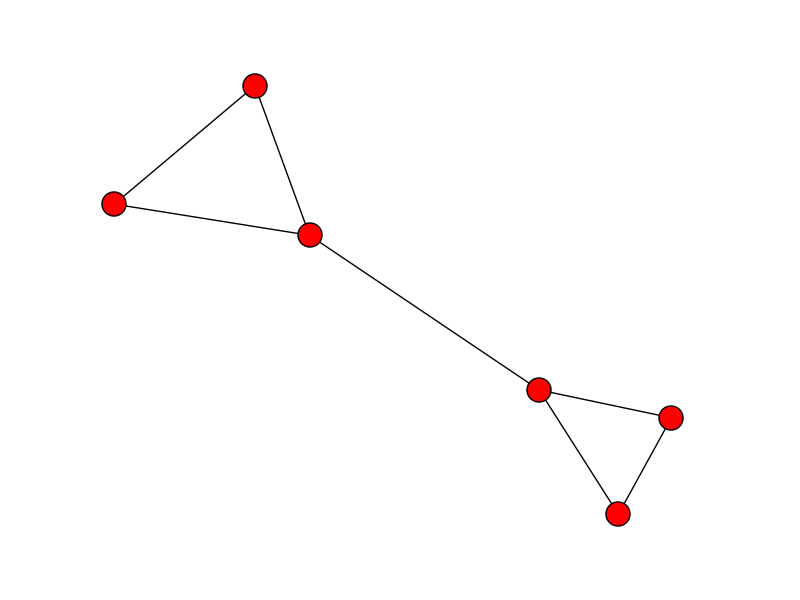
And here, the unsorted version of the points [x, y] to simulate a random points in space connected in a line:
idx = np.random.permutation(x.size)
x = x[idx]
y = y[idx]
plt.plot(x, y)
plt.show()
![enter image description here]()
The problem is then to order those points to recover their original order so that the line is plotted properly.
2. Create 2-NN graph between nodes
We can first rearrange the points in a [N, 2] array:
points = np.c_[x, y]
Then, we can start by creating a nearest neighbour graph to connect each of the nodes to its 2 nearest neighbors:
from sklearn.neighbors import NearestNeighbors
clf = NearestNeighbors(2).fit(points)
G = clf.kneighbors_graph()
G is a sparse N x N matrix, where each row represents a node, and the non-zero elements of the columns the euclidean distance to those points.
We can then use networkx to construct a graph from this sparse matrix:
import networkx as nx
T = nx.from_scipy_sparse_matrix(G)
3. Find shortest path from source
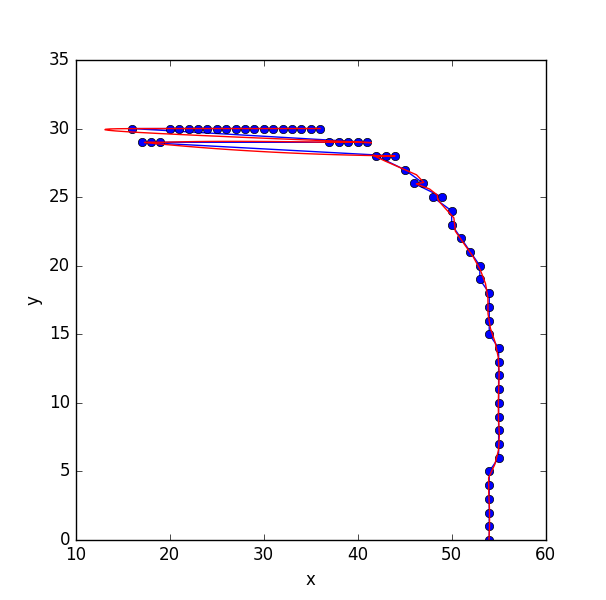
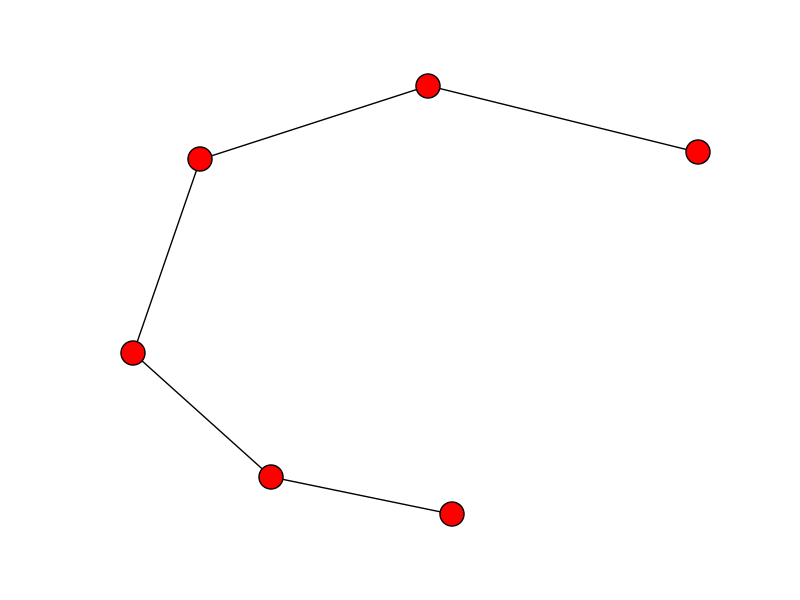
And, here begins the magic: we can extract the paths using dfs_preorder_nodes, which will essentially create a path through all the nodes (passing through each of them exactly once) given a starting node (if not given, the 0 node will be selected).
order = list(nx.dfs_preorder_nodes(T, 0))
xx = x[order]
yy = y[order]
plt.plot(xx, yy)
plt.show()
![enter image description here]()
Well, is not too bad, but we can notice that the reconstruction is not optimal. This is because the point 0 in the unordered list lays in the middle of the line, that is way it first goes in one direction, and then comes back and finishes in the other direction.
4. Find the path with smallest cost from all sources
So, in order to obtain the optimal order, we can just get the best order for all the nodes:
paths = [list(nx.dfs_preorder_nodes(T, i)) for i in range(len(points))]
Now that we have the optimal path starting from each of the N = 100 nodes, we can discard them and find the one that minimizes the distances between the connections (optimization problem):
mindist = np.inf
minidx = 0
for i in range(len(points)):
p = paths[i] # order of nodes
ordered = points[p] # ordered nodes
# find cost of that order by the sum of euclidean distances between points (i) and (i+1)
cost = (((ordered[:-1] - ordered[1:])**2).sum(1)).sum()
if cost < mindist:
mindist = cost
minidx = i
The points are ordered for each of the optimal paths, and then a cost is computed (by calculating the euclidean distance between all pairs of points i and i+1). If the path starts at the start or end point, it will have the smallest cost as all the nodes will be consecutive. On the other hand, if the path starts at a node that lies in the middle of the line, the cost will be very high at some point, as it will need to travel from the end (or beginning) of the line to the initial position to explore the other direction. The path that minimizes that cost, is the path starting in an optimal point.
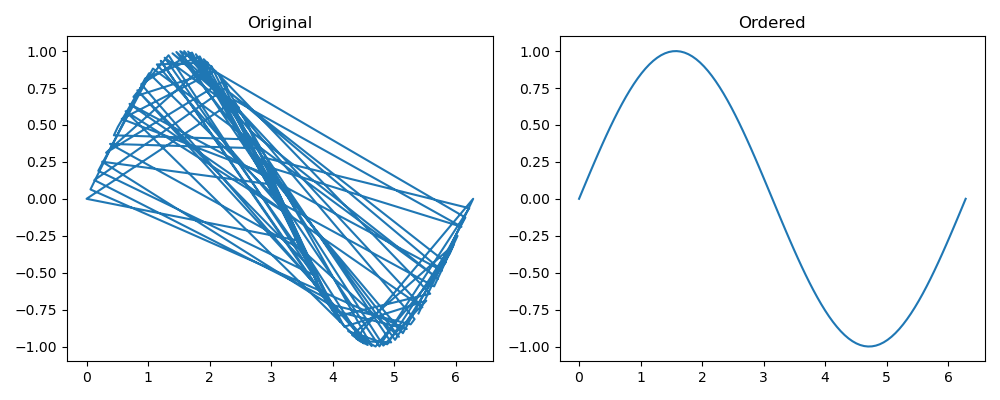
opt_order = paths[minidx]
Now, we can reconstruct the order properly:
xx = x[opt_order]
yy = y[opt_order]
plt.plot(xx, yy)
plt.show()
![enter image description here]()