Similar image search problem
- Millions of images pHash'ed and stored in Elasticsearch.
- Format is "11001101...11" (length 64), but can be changed (better not).
Given subject image's hash "100111..10" we want to find all similar image hashes in Elasticsearch index within hamming distance of 8.
Of course, query can return images with greater distance than 8 and script in Elasticsearch or outside can filter the result set. But total search time must be within 1 second or so.
Our current mapping
Each document has nested images field that contains image hashes:
{
"images": {
"type": "nested",
"properties": {
"pHashFingerprint": {"index": "not_analysed", "type": "string"}
}
}
}
Our poor solution
Fact: Elasticsearch fuzzy query supports Levenshtein distance of max 2 only.
We used custom tokenizer to split 64 bit string into 4 groups of 16 bits and do 4 group search with four fuzzy queries.
Analyzer:
{
"analysis": {
"analyzer": {
"split4_fingerprint_analyzer": {
"type": "custom",
"tokenizer": "split4_fingerprint_tokenizer"
}
},
"tokenizer": {
"split4_fingerprint_tokenizer": {
"type": "pattern",
"group": 0,
"pattern": "([01]{16})"
}
}
}
}
Then new field mapping:
"index_analyzer": "split4_fingerprint_analyzer",
Then query:
{
"query": {
"filtered": {
"query": {
"nested": {
"path": "images",
"query": {
"bool": {
"minimum_should_match": 2,
"should": [
{
"fuzzy": {
"phashFingerprint.split4": {
"value": "0010100100111001",
"fuzziness": 2
}
}
},
{
"fuzzy": {
"phashFingerprint.split4": {
"value": "1010100100111001",
"fuzziness": 2
}
}
},
{
"fuzzy": {
"phashFingerprint.split4": {
"value": "0110100100111001",
"fuzziness": 2
}
}
},
{
"fuzzy": {
"phashFingerprint.split4": {
"value": "1110100100111001",
"fuzziness": 2
}
}
}
]
}
}
}
},
"filter": {}
}
}
}
Note that we return documents that have matching images, not the images themselves, but that should not change things a lot.
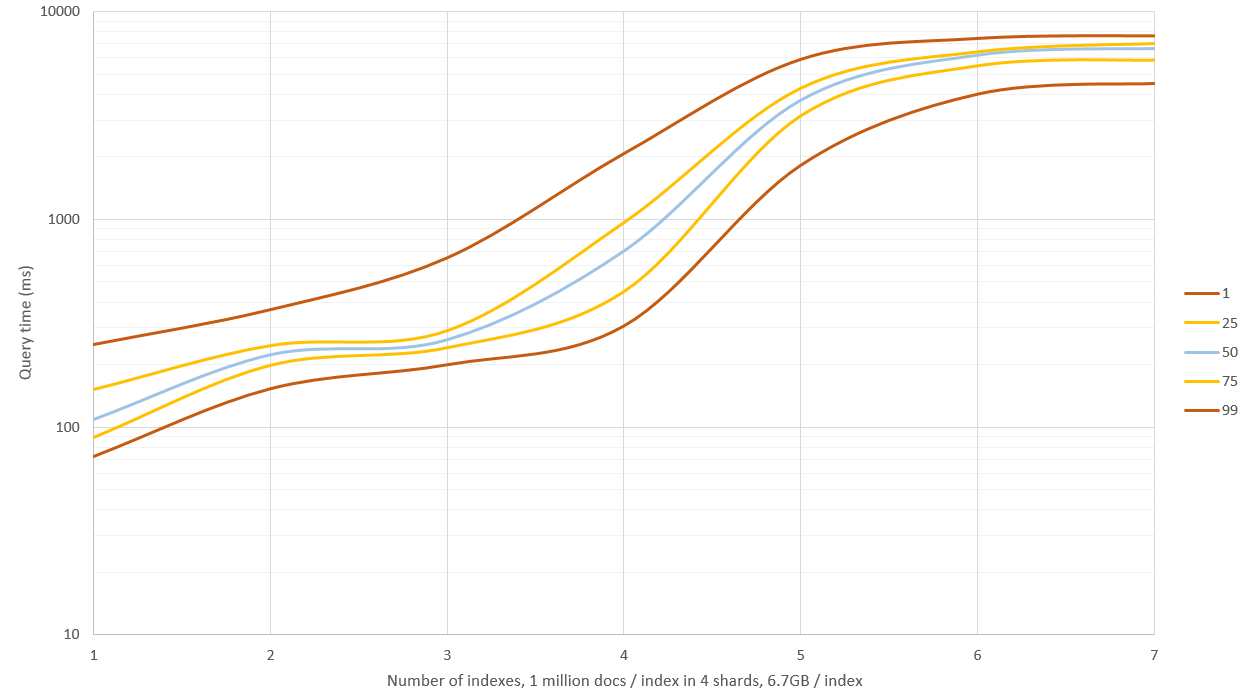
The problem is that this query returns hundreds of thousands of results even after adding other domain-specific filters to reduce initial set. Script has too much work to calculate hamming distance again, therefore query can take minutes.
As expected, if increasing minimum_should_match to 3 and 4, only subset of images that must be found are returned, but resulting set is small and fast. Below 95% of needed images are returned with minimum_should_match == 3 but we need 100% (or 99.9%) like with minimum_should_match == 2.
We tried similar approaches with n-grams, but still not much success in the similar fashion of too many results.
Any solutions of other data structures and queries?
Edit:
We noticed, that there was a bug in our evaluation process, and minimum_should_match == 2 returns 100% of results. However, processing time afterwards takes on average 5 seconds. We will see if script is worth optimising.


minimum_should_matchreturns 100%): That's great!!! If you can find a way to push your Hamming Scoring out to each Elasticsearch cluster node, you'll save on I/O and post-processing. Look atfunction_scorequeries usingscript_score. Groovy supports the XOR operator (^) and you can use Java's Integer.bitCount on the XOR output to give you Hamming Weight. – Lemonade