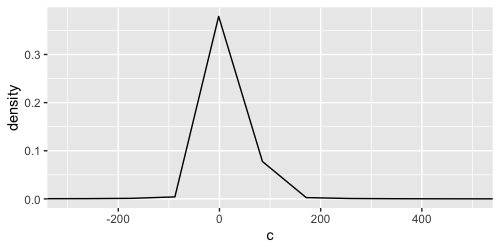
The issue is that ggplot2 by default creates 512 points along the x-axis over which to evaluate the kernel density. Since your range is fraction of the data range, only a a few of those evaluated points will be shown. We can demonstrate this by adding a point layer:
library(ggplot2)
a <- rnorm(1e4, 2, 0.5)
b <- rnorm(1e4, 0.2, 0.1)
c <- a/b
ggplot() +
geom_point(aes(c), stat = "density") +
geom_density(aes(c)) +
coord_cartesian(xlim = c(-300, 500))
![]()
Of note, density layer does not know about the coord's limits. One way to remedy the issue, is to increase the number of points with the n argument.
# Increase number of points over which to evaluate density
ggplot() +
geom_density(aes(c), n = 10000) +
coord_cartesian(xlim = c(-300, 500))
![]()
However, this can be inefficient as you're not using all these points. Instead, you can set the scale's limits, which the density layer will know about. This will make the range you're looking at more densely populated with points, giving a smoother KDE.
An important detail here is that we're using oob = scales::oob_keep as the out-of-bounds handler. This ensures that the KDE calculation considers all points, not just the ones in your range.
# Limit range to populate with points over which to evaluate density
ggplot() +
geom_density(aes(c)) +
scale_x_continuous(limits = c(-300, 500),
oob = scales::oob_keep) # Do not censor data
![]()
Created on 2023-04-24 with reprex v2.0.2




cin R, as it is also the widely used function to combine things. – Delija