Jupyter Notebook
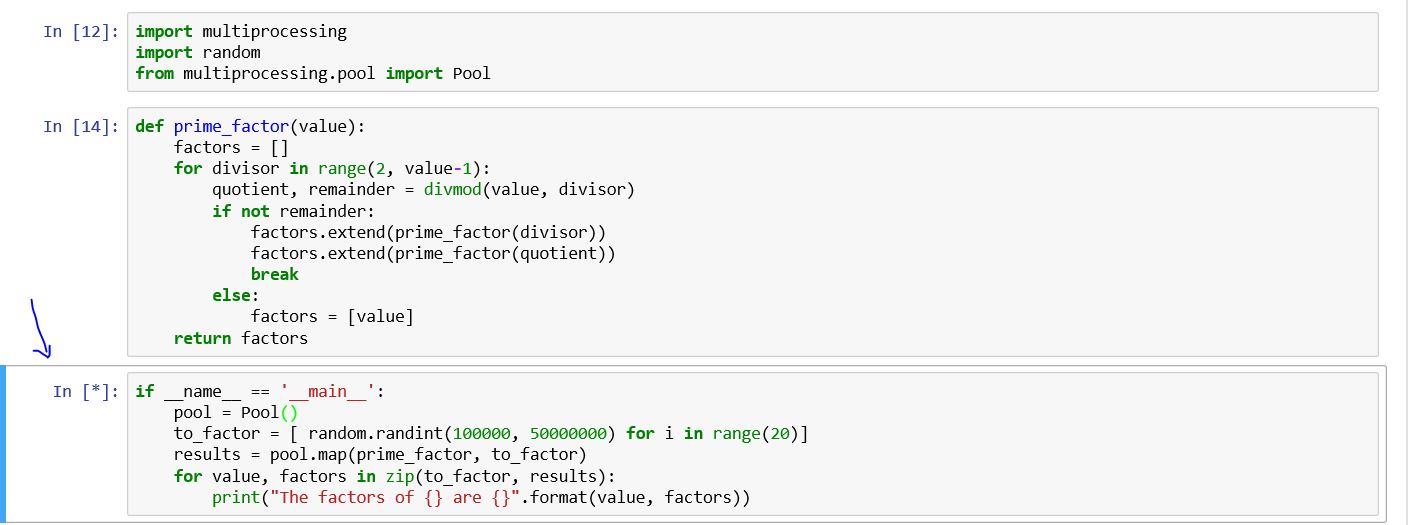
I am using multiprocessing module basically, I am still learning the capabilities of multiprocessing. I am using the book by Dusty Phillips and this code belongs to it.
import multiprocessing
import random
from multiprocessing.pool import Pool
def prime_factor(value):
factors = []
for divisor in range(2, value-1):
quotient, remainder = divmod(value, divisor)
if not remainder:
factors.extend(prime_factor(divisor))
factors.extend(prime_factor(quotient))
break
else:
factors = [value]
return factors
if __name__ == '__main__':
pool = Pool()
to_factor = [ random.randint(100000, 50000000) for i in range(20)]
results = pool.map(prime_factor, to_factor)
for value, factors in zip(to_factor, results):
print("The factors of {} are {}".format(value, factors))
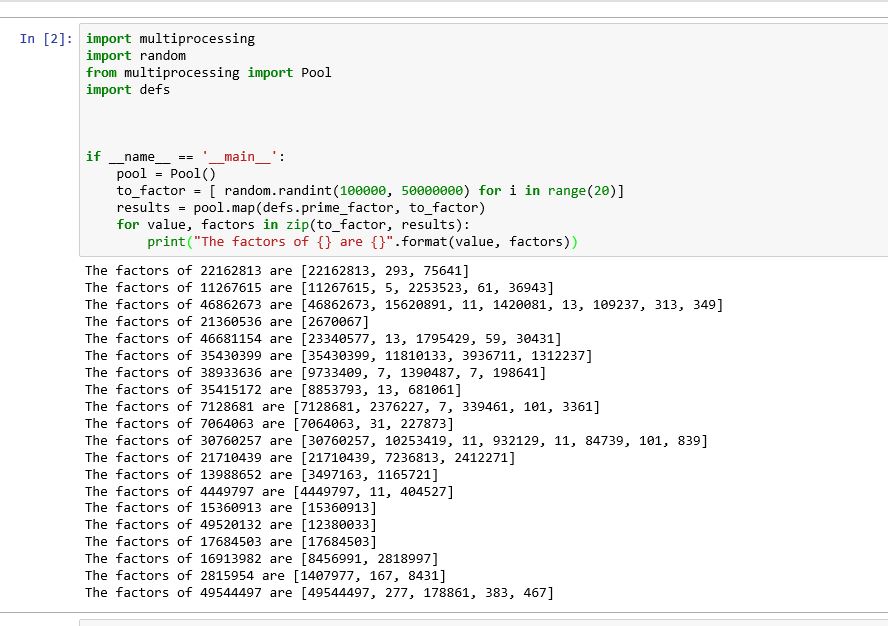
On the Windows PowerShell (not on jupyter notebook) I see the following
Process SpawnPoolWorker-5:
Process SpawnPoolWorker-1:
AttributeError: Can't get attribute 'prime_factor' on <module '__main__' (built-in)>
I do not know why the cell never ends running?