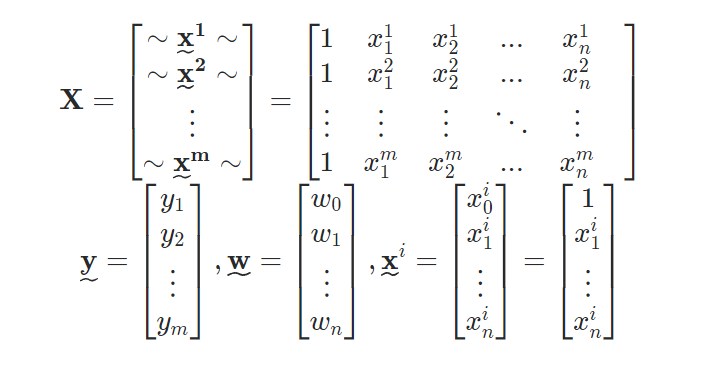
I think your code is a bit too complicated and it needs more structure, because otherwise you'll be lost in all equations and operations. In the end this regression boils down to four operations:
- Calculate the hypothesis h = X * theta
- Calculate the loss = h - y and maybe the squared cost (loss^2)/2m
- Calculate the gradient = X' * loss / m
- Update the parameters theta = theta - alpha * gradient
In your case, I guess you have confused m with n. Here m denotes the number of examples in your training set, not the number of features.
Let's have a look at my variation of your code:
import numpy as np
import random
# m denotes the number of examples here, not the number of features
def gradientDescent(x, y, theta, alpha, m, numIterations):
xTrans = x.transpose()
for i in range(0, numIterations):
hypothesis = np.dot(x, theta)
loss = hypothesis - y
# avg cost per example (the 2 in 2*m doesn't really matter here.
# But to be consistent with the gradient, I include it)
cost = np.sum(loss ** 2) / (2 * m)
print("Iteration %d | Cost: %f" % (i, cost))
# avg gradient per example
gradient = np.dot(xTrans, loss) / m
# update
theta = theta - alpha * gradient
return theta
def genData(numPoints, bias, variance):
x = np.zeros(shape=(numPoints, 2))
y = np.zeros(shape=numPoints)
# basically a straight line
for i in range(0, numPoints):
# bias feature
x[i][0] = 1
x[i][1] = i
# our target variable
y[i] = (i + bias) + random.uniform(0, 1) * variance
return x, y
# gen 100 points with a bias of 25 and 10 variance as a bit of noise
x, y = genData(100, 25, 10)
m, n = np.shape(x)
numIterations= 100000
alpha = 0.0005
theta = np.ones(n)
theta = gradientDescent(x, y, theta, alpha, m, numIterations)
print(theta)
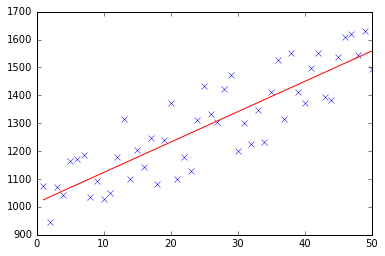
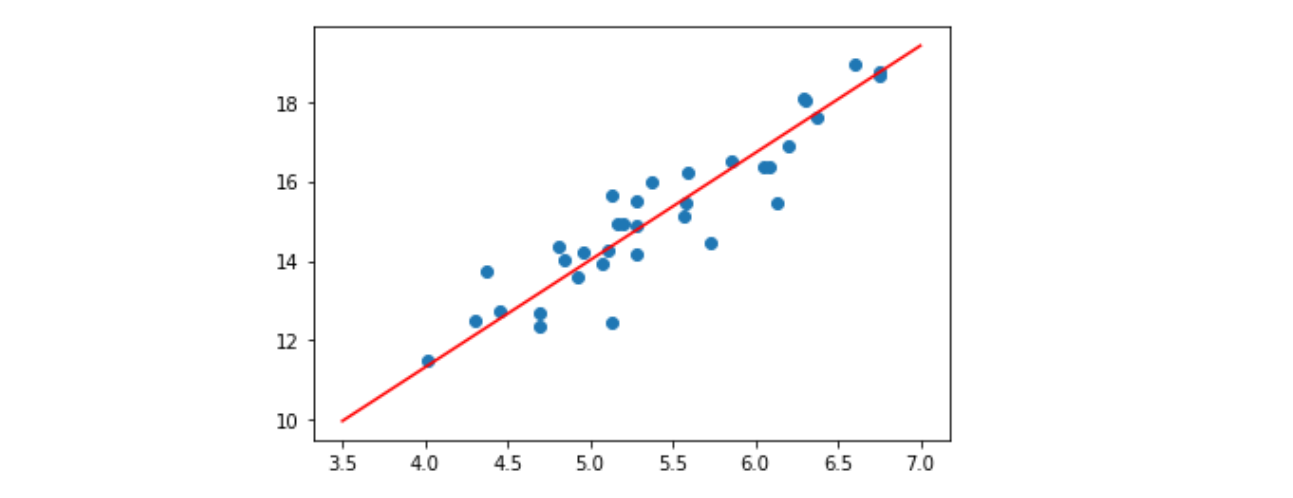
At first I create a small random dataset which should look like this:
![Linear Regression]()
As you can see I also added the generated regression line and formula that was calculated by excel.
You need to take care about the intuition of the regression using gradient descent. As you do a complete batch pass over your data X, you need to reduce the m-losses of every example to a single weight update. In this case, this is the average of the sum over the gradients, thus the division by m.
The next thing you need to take care about is to track the convergence and adjust the learning rate. For that matter you should always track your cost every iteration, maybe even plot it.
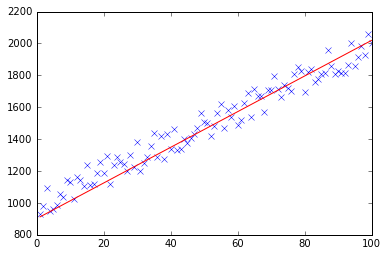
If you run my example, the theta returned will look like this:
Iteration 99997 | Cost: 47883.706462
Iteration 99998 | Cost: 47883.706462
Iteration 99999 | Cost: 47883.706462
[ 29.25567368 1.01108458]
Which is actually quite close to the equation that was calculated by excel (y = x + 30). Note that as we passed the bias into the first column, the first theta value denotes the bias weight.