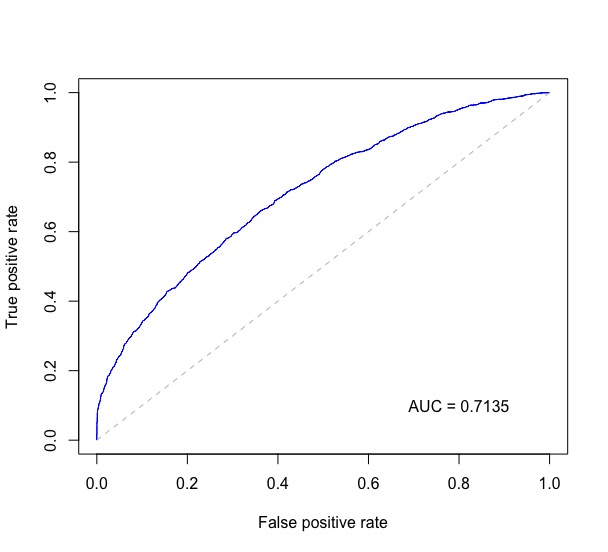
Without any additional packages:
true_Y = c(1,1,1,1,2,1,2,1,2,2)
probs = c(1,0.999,0.999,0.973,0.568,0.421,0.382,0.377,0.146,0.11)
getROC_AUC = function(probs, true_Y){
probsSort = sort(probs, decreasing = TRUE, index.return = TRUE)
val = unlist(probsSort$x)
idx = unlist(probsSort$ix)
roc_y = true_Y[idx];
stack_x = cumsum(roc_y == 2)/sum(roc_y == 2)
stack_y = cumsum(roc_y == 1)/sum(roc_y == 1)
auc = sum((stack_x[2:length(roc_y)]-stack_x[1:length(roc_y)-1])*stack_y[2:length(roc_y)])
return(list(stack_x=stack_x, stack_y=stack_y, auc=auc))
}
aList = getROC_AUC(probs, true_Y)
stack_x = unlist(aList$stack_x)
stack_y = unlist(aList$stack_y)
auc = unlist(aList$auc)
plot(stack_x, stack_y, type = "l", col = "blue", xlab = "False Positive Rate", ylab = "True Positive Rate", main = "ROC")
axis(1, seq(0.0,1.0,0.1))
axis(2, seq(0.0,1.0,0.1))
abline(h=seq(0.0,1.0,0.1), v=seq(0.0,1.0,0.1), col="gray", lty=3)
legend(0.7, 0.3, sprintf("%3.3f",auc), lty=c(1,1), lwd=c(2.5,2.5), col="blue", title = "AUC")
![enter image description here]()