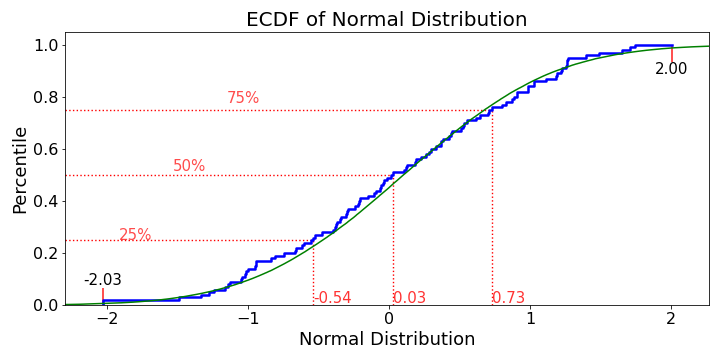
Although, there are many great answers here, though I would include a more customized ECDF plot
Generate values for the empirical cumulative distribution function
import matplotlib.pyplot as plt
def ecdf_values(x):
"""
Generate values for empirical cumulative distribution function
Params
--------
x (array or list of numeric values): distribution for ECDF
Returns
--------
x (array): x values
y (array): percentile values
"""
# Sort values and find length
x = np.sort(x)
n = len(x)
# Create percentiles
y = np.arange(1, n + 1, 1) / n
return x, y
def ecdf_plot(x, name = 'Value', plot_normal = True, log_scale=False, save=False, save_name='Default'):
"""
ECDF plot of x
Params
--------
x (array or list of numerics): distribution for ECDF
name (str): name of the distribution, used for labeling
plot_normal (bool): plot the normal distribution (from mean and std of data)
log_scale (bool): transform the scale to logarithmic
save (bool) : save/export plot
save_name (str) : filename to save the plot
Returns
--------
none, displays plot
"""
xs, ys = ecdf_values(x)
fig = plt.figure(figsize = (10, 6))
ax = plt.subplot(1, 1, 1)
plt.step(xs, ys, linewidth = 2.5, c= 'b');
plot_range = ax.get_xlim()[1] - ax.get_xlim()[0]
fig_sizex = fig.get_size_inches()[0]
data_inch = plot_range / fig_sizex
right = 0.6 * data_inch + max(xs)
gap = right - max(xs)
left = min(xs) - gap
if log_scale:
ax.set_xscale('log')
if plot_normal:
gxs, gys = ecdf_values(np.random.normal(loc = xs.mean(),
scale = xs.std(),
size = 100000))
plt.plot(gxs, gys, 'g');
plt.vlines(x=min(xs),
ymin=0,
ymax=min(ys),
color = 'b',
linewidth = 2.5)
# Add ticks
plt.xticks(size = 16)
plt.yticks(size = 16)
# Add Labels
plt.xlabel(f'{name}', size = 18)
plt.ylabel('Percentile', size = 18)
plt.vlines(x=min(xs),
ymin = min(ys),
ymax=0.065,
color = 'r',
linestyle = '-',
alpha = 0.8,
linewidth = 1.7)
plt.vlines(x=max(xs),
ymin=0.935,
ymax=max(ys),
color = 'r',
linestyle = '-',
alpha = 0.8,
linewidth = 1.7)
# Add Annotations
plt.annotate(s = f'{min(xs):.2f}',
xy = (min(xs),
0.065),
horizontalalignment = 'center',
verticalalignment = 'bottom',
size = 15)
plt.annotate(s = f'{max(xs):.2f}',
xy = (max(xs),
0.935),
horizontalalignment = 'center',
verticalalignment = 'top',
size = 15)
ps = [0.25, 0.5, 0.75]
for p in ps:
ax.set_xlim(left = left, right = right)
ax.set_ylim(bottom = 0)
value = xs[np.where(ys > p)[0][0] - 1]
pvalue = ys[np.where(ys > p)[0][0] - 1]
plt.hlines(y=p, xmin=left, xmax = value,
linestyles = ':', colors = 'r', linewidth = 1.4);
plt.vlines(x=value, ymin=0, ymax = pvalue,
linestyles = ':', colors = 'r', linewidth = 1.4)
plt.text(x = p / 3, y = p - 0.01,
transform = ax.transAxes,
s = f'{int(100*p)}%', size = 15,
color = 'r', alpha = 0.7)
plt.text(x = value, y = 0.01, size = 15,
horizontalalignment = 'left',
s = f'{value:.2f}', color = 'r', alpha = 0.8);
# fit the labels into the figure
plt.title(f'ECDF of {name}', size = 20)
plt.tight_layout()
if save:
plt.savefig(save_name + '.png')
ecdf_plot(np.random.randn(100), name='Normal Distribution', save=True, save_name="ecdf")
![enter image description here]()
Additional Resources: