I would like to contribute to this thread an algorithm that I have developed myself:
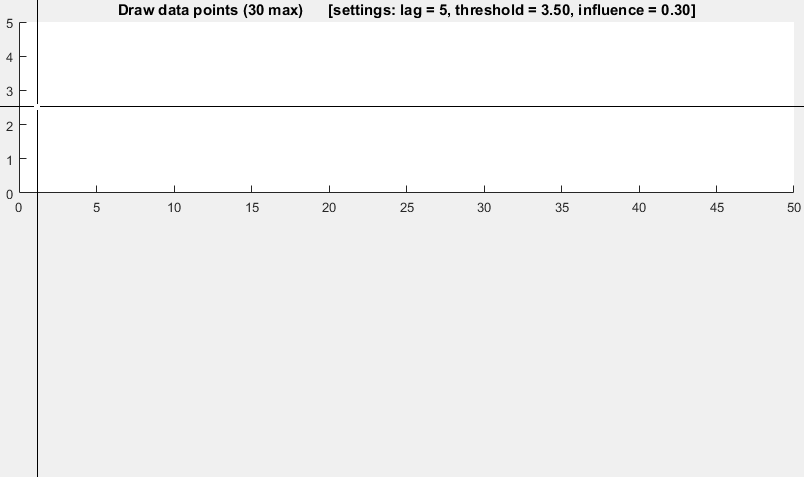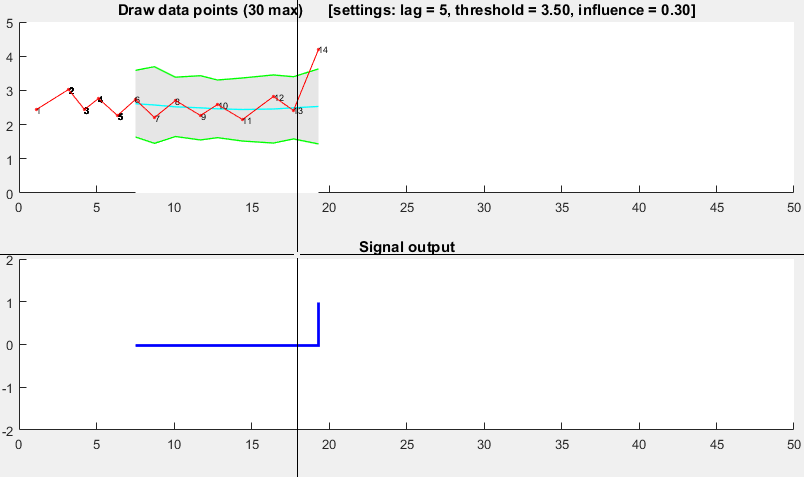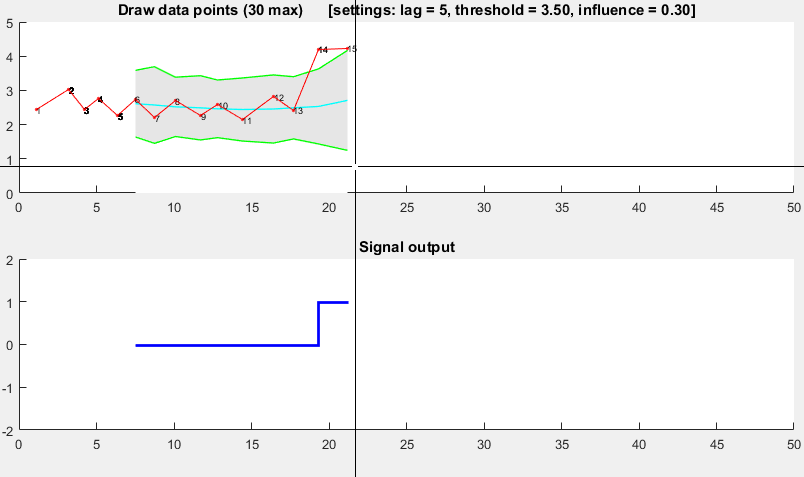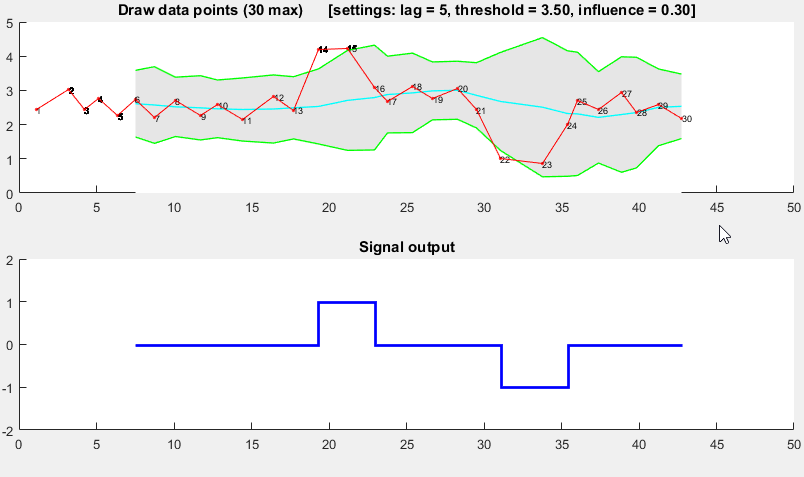
It is based on the principle of dispersion: if a new datapoint is a given x number of standard deviations away from some moving mean, the algorithm signals (also called z-score). The algorithm is very robust because it constructs a separate moving mean and deviation, such that signals do not corrupt the threshold. Future signals are therefore identified with approximately the same accuracy, regardless of the amount of previous signals. The algorithm takes 3 inputs: lag = the lag of the moving window, threshold = the z-score at which the algorithm signals and influence = the influence (between 0 and 1) of new signals on the mean and standard deviation. For example, a lag of 5 will use the last 5 observations to smooth the data. A threshold of 3.5 will signal if a datapoint is 3.5 standard deviations away from the moving mean. And an influence of 0.5 gives signals half of the influence that normal datapoints have. Likewise, an influence of 0 ignores signals completely for recalculating the new threshold: an influence of 0 is therefore the most robust option.
It works as follows:
Pseudocode
# Let y be a vector of timeseries data of at least length lag+2
# Let mean() be a function that calculates the mean
# Let std() be a function that calculates the standard deviaton
# Let absolute() be the absolute value function
# Settings (the ones below are examples: choose what is best for your data)
set lag to 5; # lag 5 for the smoothing functions
set threshold to 3.5; # 3.5 standard deviations for signal
set influence to 0.5; # between 0 and 1, where 1 is normal influence, 0.5 is half
# Initialise variables
set signals to vector 0,...,0 of length of y; # Initialise signal results
set filteredY to y(1,...,lag) # Initialise filtered series
set avgFilter to null; # Initialise average filter
set stdFilter to null; # Initialise std. filter
set avgFilter(lag) to mean(y(1,...,lag)); # Initialise first value
set stdFilter(lag) to std(y(1,...,lag)); # Initialise first value
for i=lag+1,...,t do
if absolute(y(i) - avgFilter(i-1)) > threshold*stdFilter(i-1) then
if y(i) > avgFilter(i-1)
set signals(i) to +1; # Positive signal
else
set signals(i) to -1; # Negative signal
end
# Adjust the filters
set filteredY(i) to influence*y(i) + (1-influence)*filteredY(i-1);
set avgFilter(i) to mean(filteredY(i-lag,i),lag);
set stdFilter(i) to std(filteredY(i-lag,i),lag);
else
set signals(i) to 0; # No signal
# Adjust the filters
set filteredY(i) to y(i);
set avgFilter(i) to mean(filteredY(i-lag,i),lag);
set stdFilter(i) to std(filteredY(i-lag,i),lag);
end
end
Demo
![Demonstration of robust thresholding algorithm]()