Another possible solution.
I'll create some random data.
import numpy as np
import pandas as pd
import scipy.stats as sps
from sklearn import linear_model
from sklearn.metrics import roc_curve, RocCurveDisplay, auc
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
# define data distributions
N0 = 300
N1 = 250
dist0 = sps.gamma(a=8, scale=1/10)
x0 = np.linspace(dist0.ppf(0), dist0.ppf(1-1e-5), 100)
y0 = dist0.pdf(x0)
dist1 = sps.gamma(a=15, scale=1/10)
x1 = np.linspace(dist1.ppf(0), dist1.ppf(1-1e-5), 100)
y1 = dist1.pdf(x1)
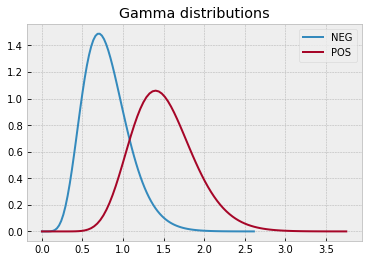
with plt.style.context("bmh"):
plt.plot(x0, y0, label="NEG")
plt.plot(x1, y1, label="POS")
plt.legend()
plt.title("Gamma distributions")
![enter image description here]()
# create a random dataset
rvs0 = dist0.rvs(N0, random_state=0)
rvs1 = dist1.rvs(N1, random_state=1)
with plt.style.context("bmh"):
plt.hist(rvs0, alpha=.5, label="NEG")
plt.hist(rvs1, alpha=.5, label="POS")
plt.legend()
plt.title("Random dataset")
![enter image description here]()
Initialize a dataframe with observations (x feature and y target)
df = pd.DataFrame({
"y": np.concatenate(( np.repeat(0, N0) , np.repeat(1, N1) )),
"x": np.concatenate(( rvs0 , rvs1 )),
})
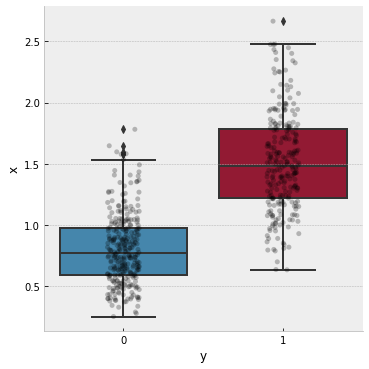
and display it with a box plot
# plot the data
with plt.style.context("bmh"):
g = sns.catplot(
kind="box",
data=df,
x="y", y="x"
)
ax = g.axes.flat[0]
sns.stripplot(
data=df,
x="y", y="x",
ax=ax, color='k',
alpha=.25
)
plt.show()
![enter image description here]()
Now, we can split the dataframe into train-test, perform Logistic regression, compute ROC curve, AUC, Youden's index, find the cut-off and plot everything. All using pandas
# split dataset into train-test
X_train, X_test, y_train, y_test = train_test_split(
df[["x"]], df.y.values, test_size=0.5, random_state=1)
# init and fit Logistic Regression on train set
clf = linear_model.LogisticRegression()
clf.fit(X_train, y_train)
# predict probabilities on x test set
y_proba = clf.predict_proba(X_test)
# compute FPR and TPR from y test set and predicted probabilities
fpr, tpr, thresholds = roc_curve(
y_test, y_proba[:,1], drop_intermediate=False)
# compute ROC AUC
roc_auc = auc(fpr, tpr)
# init a dataframe for results
df_test = pd.DataFrame({
"x": X_test.x.values.flatten(),
"y": y_test,
"proba": y_proba[:,1]
})
# sort it by predicted probabilities
# because thresholds[1:] = y_proba[::-1]
df_test.sort_values(by="proba", inplace=True)
# add reversed TPR and FPR
df_test["tpr"] = tpr[1:][::-1]
df_test["fpr"] = fpr[1:][::-1]
# optional: add thresholds to check
#df_test["thresholds"] = thresholds[1:][::-1]
# add Youden's j index
df_test["youden_j"] = df_test.tpr - df_test.fpr
# define the cut_off and diplay it
cut_off = df_test.sort_values(
by="youden_j", ascending=False, ignore_index=True).iloc[0]
print("CUT-OFF:")
print(cut_off)
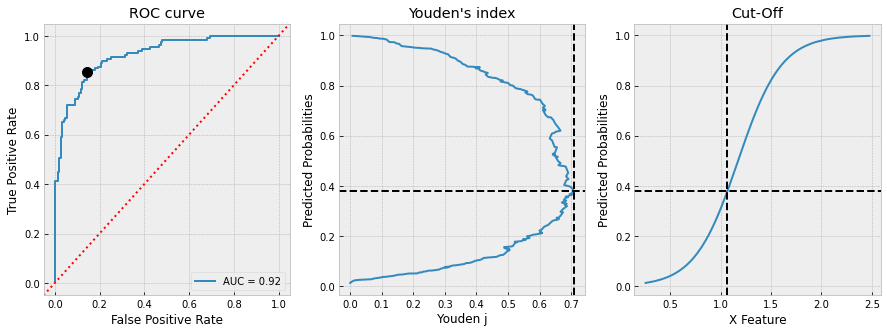
# plot everything
with plt.style.context("bmh"):
fig, ax = plt.subplots(1, 3, figsize=(15, 5))
RocCurveDisplay(
fpr=df_test.fpr, tpr=df_test.tpr,
roc_auc=roc_auc).plot(ax=ax[0])
ax[0].set_title("ROC curve")
ax[0].axline(xy1=(0,0), slope=1, color="r", ls=":")
ax[0].plot(cut_off.fpr, cut_off.tpr, 'ko', ms=10)
df_test.plot(
x="youden_j", y="proba", ax=ax[1],
ylabel="Predicted Probabilities", xlabel="Youden j",
title="Youden's index", legend=False
)
ax[1].axvline(cut_off.youden_j, color="k", ls="--")
ax[1].axhline(cut_off.proba, color="k", ls="--")
df_test.plot(
x="x", y="proba", ax=ax[2],
ylabel="Predicted Probabilities", xlabel="X Feature",
title="Cut-Off", legend=False
)
ax[2].axvline(cut_off.x, color="k", ls="--")
ax[2].axhline(cut_off.proba, color="k", ls="--")
plt.show()
and we get
CUT-OFF:
x 1.065712
y 1.000000
proba 0.378543
tpr 0.852713
fpr 0.143836
youden_j 0.708878
![enter image description here]()
We can finally check
# check results
TP = df_test[(df_test.x>=cut_off.x)&(df_test.y==1)].index.size
FP = df_test[(df_test.x>=cut_off.x)&(df_test.y==0)].index.size
TN = df_test[(df_test.x< cut_off.x)&(df_test.y==0)].index.size
FN = df_test[(df_test.x< cut_off.x)&(df_test.y==1)].index.size
print("True Positive Rate: ", TP / (TP + FN))
print("False Positive Rate:", 1 - TN / (TN + FP))
True Positive Rate: 0.8527131782945736
False Positive Rate: 0.14383561643835618


thresholds[np.argmax(tpr - fpr)]and the most upvoted answer functionthreshold = Find_Optimal_Cutoff(data['true'], data['pred'])? The thresholds are close, but different when I do an actual calculation. – Wiviniathresholds[np.argmin(abs(tpr-(1-fpr)))]– Josefajosefinathresholds[np.argmin((1 - tpr) ** 2 + fpr ** 2)]. But @cgnorthcutt's solution maximizes the Youden's J statistic, which seems to be the more accepted method. What is truly "optimal" for your situation depends on the relative costs of false positives and false negatives. – Selfaddressed