@Ben's solution is pretty sweet. It doesn't handle the follwing cases though:
# all these return numeric(0):
x <- c(1,2,9,9,2,1,1,5,5,1) # duplicated points at maxima
which(diff(sign(diff(x)))==-2)+1
x <- c(2,2,9,9,2,1,1,5,5,1) # duplicated points at start
which(diff(sign(diff(x)))==-2)+1
x <- c(3,2,9,9,2,1,1,5,5,1) # start is maxima
which(diff(sign(diff(x)))==-2)+1
Here's a more robust (and slower, uglier) version:
localMaxima <- function(x) {
# Use -Inf instead if x is numeric (non-integer)
y <- diff(c(-.Machine$integer.max, x)) > 0L
rle(y)$lengths
y <- cumsum(rle(y)$lengths)
y <- y[seq.int(1L, length(y), 2L)]
if (x[[1]] == x[[2]]) {
y <- y[-1]
}
y
}
x <- c(1,2,9,9,2,1,1,5,5,1)
localMaxima(x) # 3, 8
x <- c(2,2,9,9,2,1,1,5,5,1)
localMaxima(x) # 3, 8
x <- c(3,2,9,9,2,1,1,5,5,1)
localMaxima(x) # 1, 3, 8


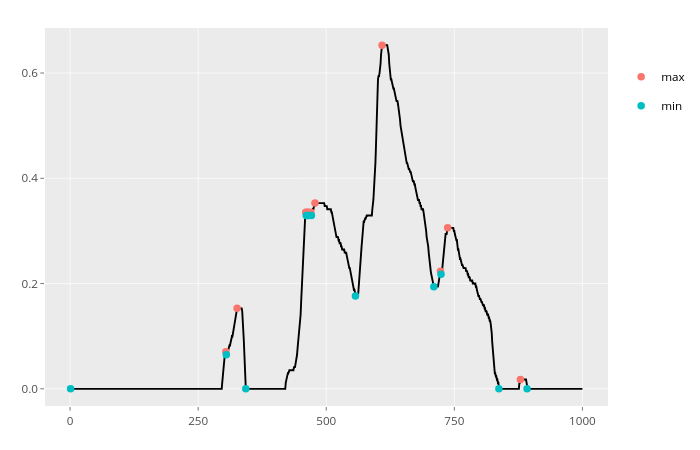
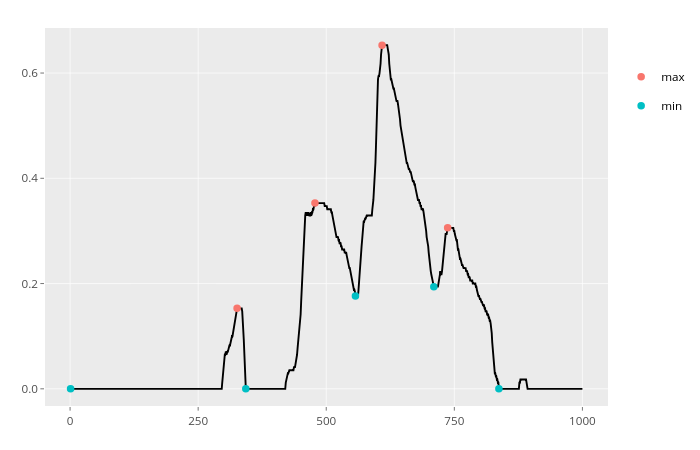
which(diff(sign(diff(x)))==-2)+1if the values aren't always changing by one. – Spicate