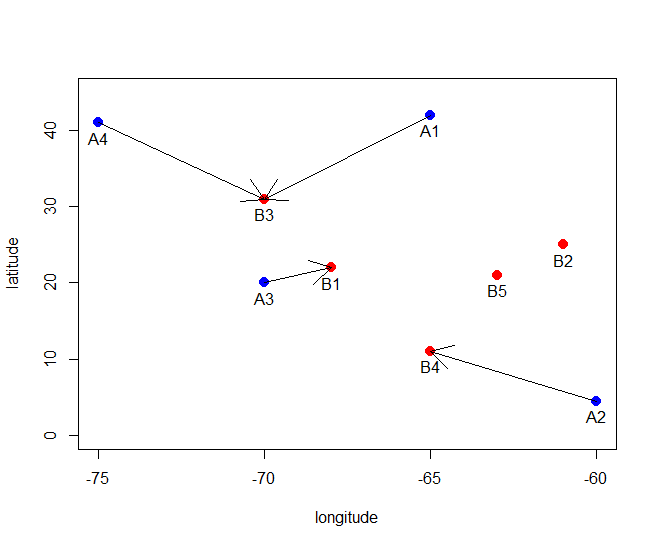
I am working on a study that is trying to assign particulate matter exposure to specific individuals based on their addresses. I have two data sets with longitude and latitude coordinates. One if for individuals and one if for pm exposure blocks. I want to assign each subject with a pm exposure block based on the block that is closest.
library(sp)
library(raster)
library(tidyverse)
#subject level data
subjectID<-c("A1","A2","A3","A4")
subjects<-data.frame(tribble(
~lon,~lat,
-70.9821391, 42.3769511,
-61.8668537, 45.5267133,
-70.9344039, 41.6220337,
-70.7283830, 41.7123494
))
row.names(subjects)<-subjectID
#PM Block Locations
blockID<-c("B1","B2","B3","B4","B5")
blocks<-data.frame(tribble(
~lon,~lat,
-70.9824591, 42.3769451,
-61.8664537, 45.5267453,
-70.9344539, 41.6220457,
-70.7284530, 41.7123454,
-70.7284430, 41.7193454
))
row.names(blocks)<-blockID
#Creating distance matrix
dis_matrix<-pointDistance(blocks,subjects,lonlat = TRUE)
###The above code doesnt preserve the row names. Is there a way to to do
that?
###I'm unsure about the below code
colnames(dis_matrix)<-row.names(subjects)
row.names(dis_matrix)<-row.names(blocks)
dis_data<-data.frame(dis_matrix)
###Finding nearst neighbor and coercing to usable format
getname <-function(x) {
row.names(dis_data[which.min(x),])
}
nn<-data.frame(lapply(dis_data,getname)) %>%
gather(key=subject,value=neighbor)
This code gives me output that makes sense but I'm unsure of the validity and efficiency. Any suggestion on how to improve and fix this code are appreciated. I also receive the error message:
Warning message:
attributes are not identical across measure variables;
they will be dropped
Which I can't determine the origin of.
Thanks for taking a look!