I have a graph with 480k nodes and 34M edges. I want to create node embeddings using Node2Vec on this graph. But, It is not even able to calculate transition probabilities. I am using a Google Cloud Machine with 32 cores and 120 GB RAM. Infrastructure is not the problem, the problem is that the function _precompute_probabilities in the node2vec pip library is not paraller. It is using only a single thread to calculate the transition probabilities. Is there a way to make this parallel or is they any other parallel version of Node2Vec ?
TLDR
To compute embeddings on large graphs use GRAPE.
pip install grape
Example:
from grape import Graph
from grape.embedders import Node2VecGloVeEnsmallen
graph = Graph.from_csv(
## The path to the edges list tsv
edge_path="edges.csv",
sources_column="source",
destinations_column="destination",
directed=False,
)
embedding = Node2VecGloVeEnsmallen().fit_transform(graph)
Longer answer
To solve this issue, we developed GRAPE, our Rust library with Python bindings. We needed to run node2vec on big graphs and the libraries we found weren't fast enough. We re-implemented and optimized many models, including Node2vec's, from the ground up without using Tensorflow or Pytorch.
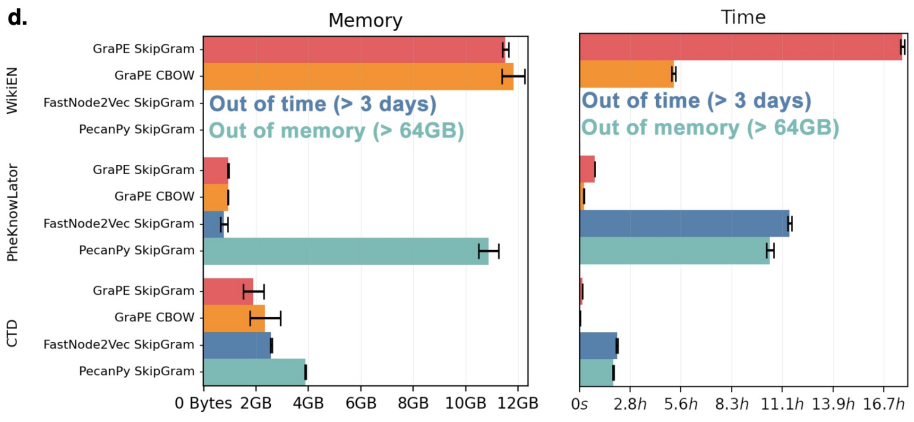
Here's some benchmarks on our server with 12 cores (24 threads) and 128GB of ram.
- WikiEN has 130M edges and 17M nodes.
- CTD has 45M edges and 100K nodes.
- PheKnowLator has 7M edges and 800K nodes.
Here's the tutorial on how to load your own custom graph, we support CSV-like formats: https://github.com/AnacletoLAB/grape/blob/main/tutorials/Loading_a_Graph_in_Ensmallen.ipynb
and here's the complete tutorial on how to run and visualize node2vec with Glove on a given graph:
https://github.com/AnacletoLAB/grape/blob/main/tutorials/Using_Node2Vec_GloVe_to_embed_Cora.ipynb
Our paper is still under review ( https://arxiv.org/abs/2110.06196 ) and we are just two developers, so, if you need any help contact us on Discord, Github, or Twitter @GRAPElib.
from grape import GraphBuilder and then you can use add_node and add_edge to manually compose the graph. Alternatively, you can load a pandas dataframe –
Ranna I found a library Graph2Vec, it uses a CSR Matrix to generate walks instead of jumping from node to node in memory. It is way faster than Node2Vec.
Link: https://www.singlelunch.com/2019/08/01/700x-faster-node2vec-models-fastest-random-walks-on-a-graph/
Github: https://github.com/VHRanger/graph2vec
Also, you can refer to this issue and try the mentioned libraries: https://github.com/aditya-grover/node2vec/issues/10
I've tried https://github.com/eliorc/node2vec with the "temp_folder" property. Thought I didn't feel that it was much faster, so I ended up with the version with CSR Matrices. Oh... was it yourself, who answered the question? :) Good to know, thank you for the tip
© 2022 - 2024 — McMap. All rights reserved.