Suppose I have this dataset (the actual dataset has 30+ columns and thousands of ids)
df <- data.frame(id = 1:5,
admission = c("Severe", "Mild", "Mild", "Moderate", "Severe"),
d1 = c(NA, "Moderate", "Mild", "Moderate", "Severe"),
d2 = c(NA, "Moderate", NA, "Mild", "Moderate"),
d3 = c(NA, "Severe", NA, "Mild", NA),
d4 = c(NA, NA, NA, "Mild", NA),
outcome = c("Dead", "Dead", "Alive", "Alive", "Dead"))
I want to make a Sankey diagram that illustrates the daily severity of the patients over time. However, when the observation reaches NA (means that an outcome has been reached), I want the node to directly link to the outcome.
This is how the diagram should look like:
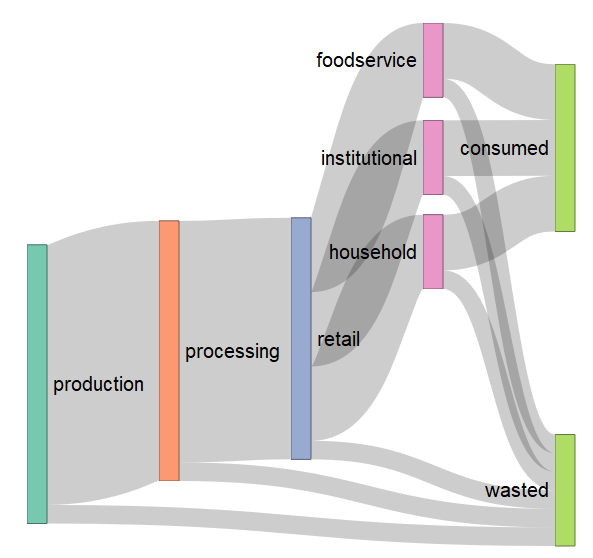
Image fetched from the question asked by @qdread here
Is this possible with ggsankey?
This is my current code:
df.sankey <- df %>%
make_long(admission, d1, d2, d3, d4, outcome)
ggplot(df.sankey, aes(x = x,
next_x = next_x,
node = node,
next_node = next_node,
fill = factor(node),
label = node)) +
geom_sankey(flow. Alpha = 0.5,
node. Color = NA,
show. Legend = TRUE) +
geom_sankey_text(size = 3, color = "black", fill = NA, hjust = 0, position = position_nudge(x = 0.1))
EDIT
Based on the solution provided by @Allan Cameron, I managed to bypass the nodes with NA values. However, the diagram looks quite complex because the links to the targets are not sorted.
do.call(rbind, apply(df, 1, function(x) {
x <- na.omit(x[-1])
data.frame(x = names(x), node = x,
next_x = dplyr::lead(names(x)),
next_node = dplyr::lead(x), row.names = NULL)
})) %>%
ggplot(df.sankey, aes(x = x,
next_x = next_x,
node = node,
next_node = next_node,
fill = factor(node),
label = node)) +
geom_sankey(flow.alpha = 0.5,
node.color = NA,
show.legend = TRUE) +
geom_sankey_text(size = 3, color = "black", fill = NA, hjust = 0, position = position_nudge(x = 0.1))
which results in this diagram:
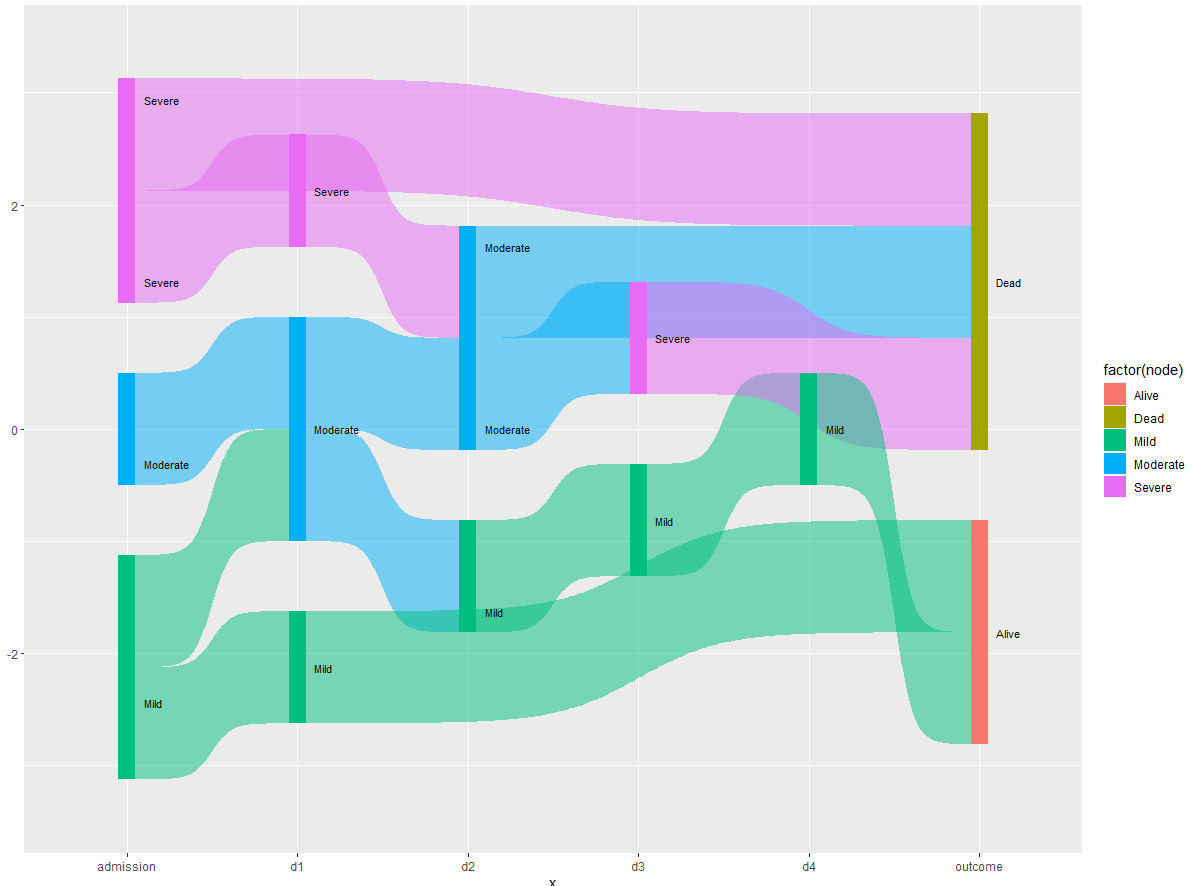
Is it possible to sort the links to the Outcome target so that all links with Severe value gets aggregated?
Thanks in advance for the help.

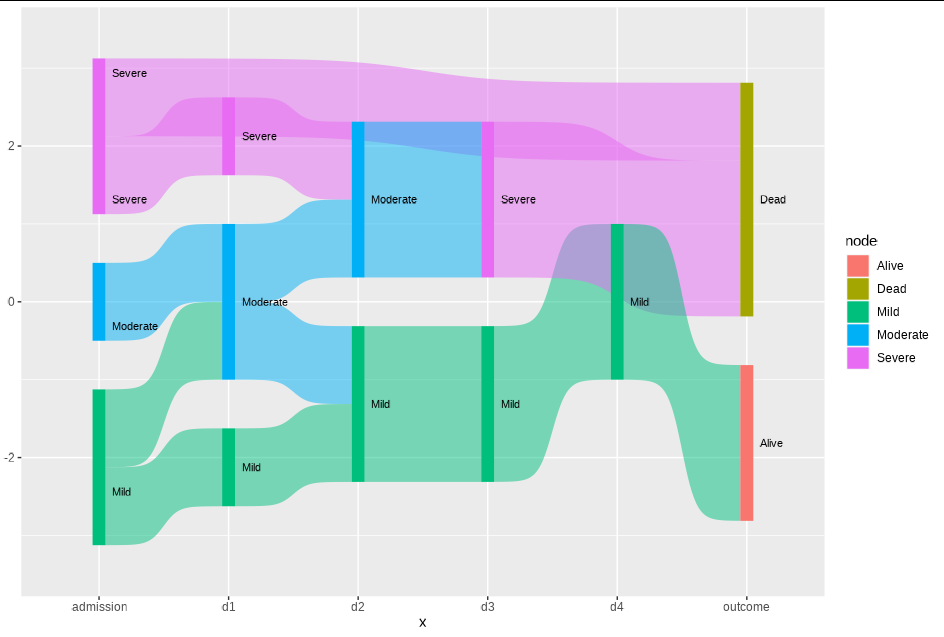
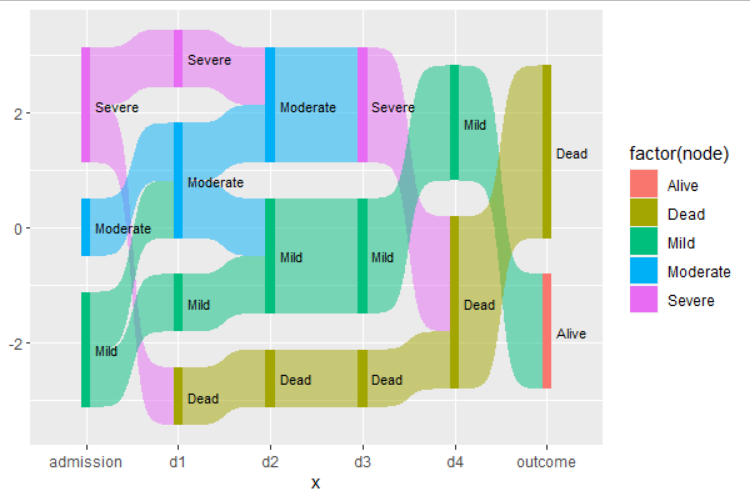
Outcomevertical bar? Since the real dataset has tens of columns, the branches looks too complex as they are not sorted – Post