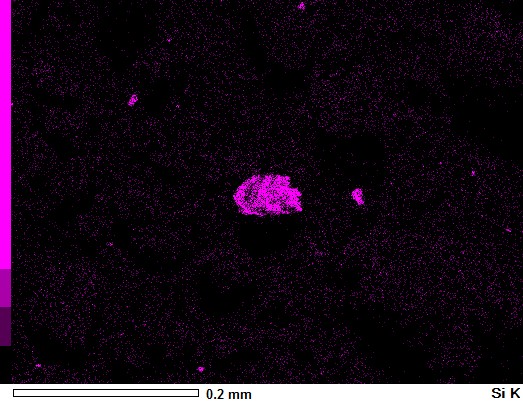
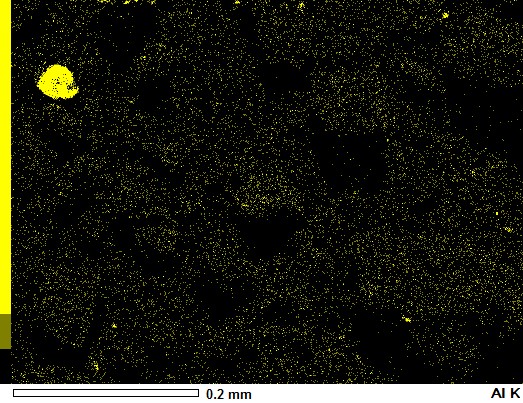
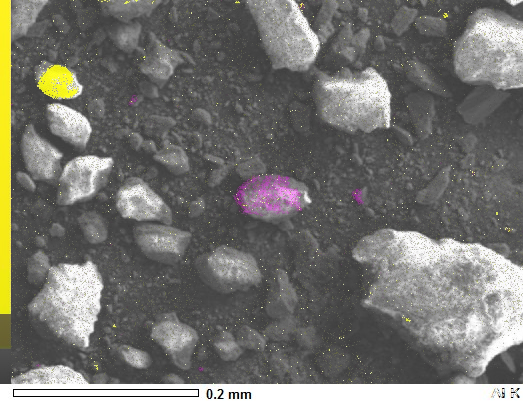
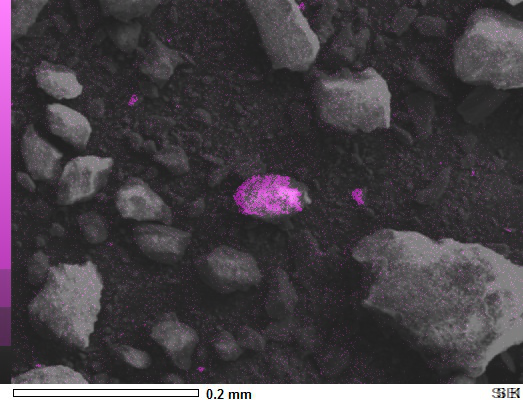
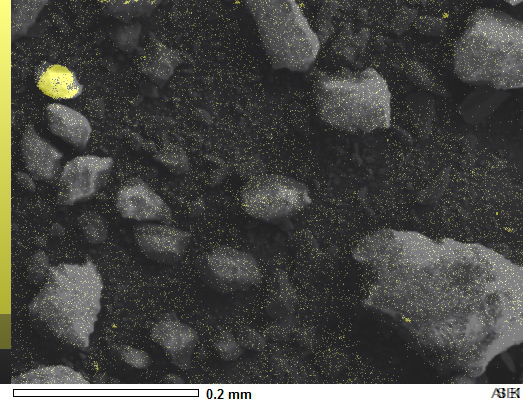
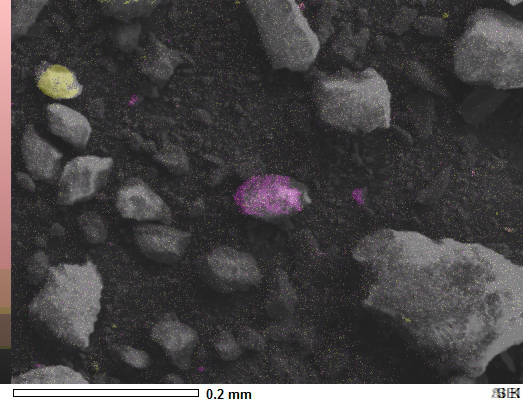
You don't need Numpy arrays at all for a simple overlay.
from PIL import Image
# Change to your file names, this is what mine downloaded as from your post
image_sem = Image.open('aZ8ED.jpg')
image_si = Image.open('HeiA7.jpg')
image_al = Image.open('HlcOR.jpg')
si_k = Image.blend(image_sem, image_si, 0.5)
si_k.show()
al_k = Image.blend(image_sem, image_al, 0.5)
al_k.show()
You'll have to crop or work with the picture label in the lower right corner for your needs and perhaps the color scale on the left, but this should get you started. It worked for me.
![enter image description here]()
![enter image description here]()
Edit based on OP comments:
The blend method outputs out = image1 * (1.0 - alpha) + image2 * alpha. To put all of them together, just successively combine the element images together, then blend that result with the master image as follows:
elements = Image.blend(image_si, image_al, 0.5)
elements_overlay = Image.blend(image_sem, elements, 0.5)
elements_overlay.show()
![enter image description here]()
The blend method may not be the best for many images. The colors will fade, as the alpha makes the first element image a smaller weight of the final image as more element images are combined. See documentation for all means of combining images. For more complex combinations, you may want to use the Numpy array after all and do some normalization or tweaking of the actual pixels then recombine with fromarray or similar.