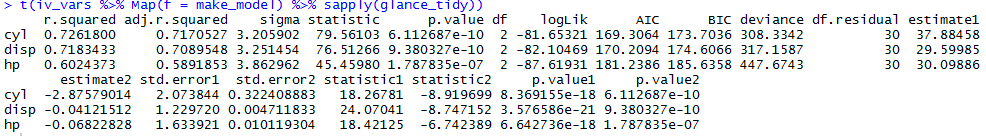
You could cumulatively paste over your vector of id_vars to get the combinations you want. I used the code in this answer to do this.
I use the plus sign as the separator between variables to get ready for the formula notation in lm.
cumpaste = function(x, .sep = " ") {
Reduce(function(x1, x2) paste(x1, x2, sep = .sep), x, accumulate = TRUE)
}
( iv_vars_cum = cumpaste(iv_vars, " + ") )
[1] "cyl" "cyl + disp" "cyl + disp + hp"
Then switch the make_model function to use a formula and a dataset. The explanatory variables, separated by the plus sign, get passed to the function after the tilde in the formula. Everything is pasted together, which lm conveniently interprets as a formula.
make_model = function(nm) {
lm(paste0("mpg ~", nm), data = mtcars)
}
Which we can see works as desired, returning a model with both explanatory variables.
make_model("cyl + disp")
Call:
lm(formula = as.formula(paste0("mpg ~", nm)), data = mtcars)
Coefficients:
(Intercept) cyl disp
34.66099 -1.58728 -0.02058
You'll likely need to rethink how you want to combine the info together, as you will now how differing numbers of columns due to the increased number of coefficients.
A possible option is to add dplyr::bind_rows to your glance_tidy function and then use map_dfr from purrr for the final output.
glance_tidy = function(x) {
dplyr::bind_rows( c( unlist(glance(x)), unlist(tidy(x)[, -1]) ) )
}
iv_vars_cum %>%
Map(f = make_model) %>%
map_dfr(glance_tidy, .id = "model")
# A tibble: 3 x 28
model r.squared adj.r.squared sigma statistic p.value df logLik AIC
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 cyl 0.7261800 0.7170527 3.205902 79.56103 6.112687e-10 2 -81.65321 169.3064
2 cyl + disp 0.7595658 0.7429841 3.055466 45.80755 1.057904e-09 3 -79.57282 167.1456
3 cyl + disp + hp 0.7678877 0.7430186 3.055261 30.87710 5.053802e-09 4 -79.00921 168.0184 ...