Reproducible Data:
data(crabs, package = "MASS")
df <- crabs[-(1:3)]
set.seed(12345)
df$GRP <- kmeans(df, 4)$cluster
df.order <- dplyr::arrange(df, GRP)
Data Description:
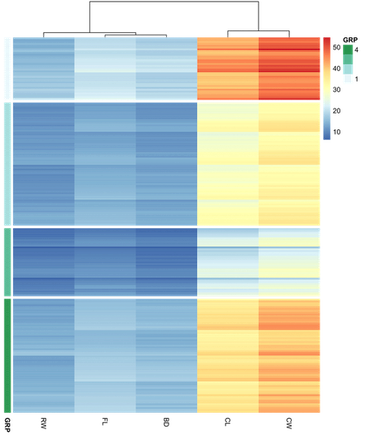
df has 5 numerical variables. I did the K-means algorithm according to these 5 attributes and produced a new categorical variable GRP which has 4 levels. Next, I ordered it with GRP and named it df.order.
What I did with pheatmap:
## 5 numerical variables for coloring
colormat <- df.order[c("FL", "RW", "CL", "CW", "BD")]
## Specify the annotation variable `GRP` shown on left side of the heatmap
ann_row <- df.order["GRP"]
## gap indices
gapRow <- cumsum(table(ann_row$GRP))
library(pheatmap)
pheatmap(colormat, cluster_rows = F, show_rownames = F,
annotation_row = ann_row, gaps_row = gapRow)
Error in annotation_colors[[colnames(annotation)[i]]] : subscript out of bounds
Here is where I got something weird:
At first, I guess the problem resulted from the argument annotation_row.I check the row names of the two data frames.
all.equal(rownames(colormat), rownames(ann_row))
# [1] TRUE
You can see that they are equal. However, I executed the following code and the heatmap work.
rownames(colormat) <- rownames(ann_row)
pheatmap(colormat, cluster_rows = F, show_rownames = F,
annotation_row = ann_row, gaps_row = gapRow)
Theoretically this code "rownames(colormat) <- rownames(ann_row)" should make no sense because these two objects are equal originally, but why does it make the pheatmap() function work?
Edit: From @steveb's comment, I don't even have to set the rownames using ann_row. I just set
rownames(colormat) <- rownames(colormat)
and the pheatmap also works. This situation is still counterintuitive.
Final Output:

rownames(colormat) <- rownames(colormat), thenpheatmapwill work; you don't even have to set therownamesusingann_row. – Parquet