I am building a binomial regression model using 2 categorical variables. This is from an example in the book, Statistical rethinking. In the book, while using the rethinking package, we can set priors on each categorical variable as shown below
m11.5 <- ulam(
alist(
pulled_left ~ dbinom( 1 , p ) ,
logit(p) <- a[actor] + b[treatment] ,
a[actor] ~ dnorm( 0 , 1.5 ),
b[treatment] ~ dnorm( 0 , 0.5 )
) ,
data=d , chains=4 , log_lik=TRUE )
I am trying to do the same in brms.
priors <- c(prior(normal(0, 1.5), class = b, coef = "actor"),
prior(normal(0, 0.5), class = b, coef = "treatment"))
m11.5 <- brm(data = d, family = binomial,
formula = pulled_left | trials(1) ~ 1 + actor + treatment,
sample_prior = T, prior = priors,
cores = 4, chains = 4)
I would like to set priors for all of the actor levels and the treatment levels mentioned once. However the above code doesn't go through with the following error message,
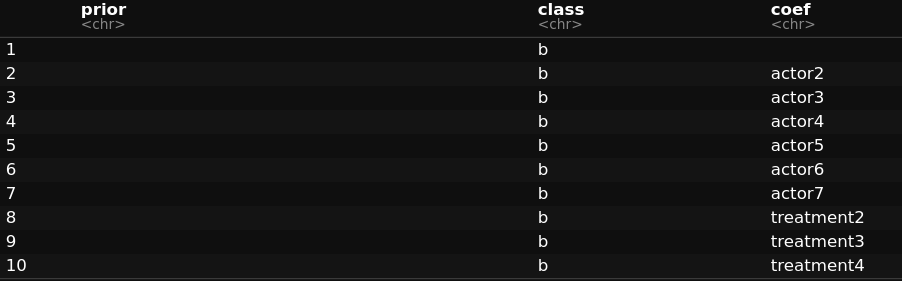
Upon using get_prior, I see the following (implying that the levels are internally split)
I donot want to specify the prior for the each level of the categorical variables. How do I do it? Please advice.