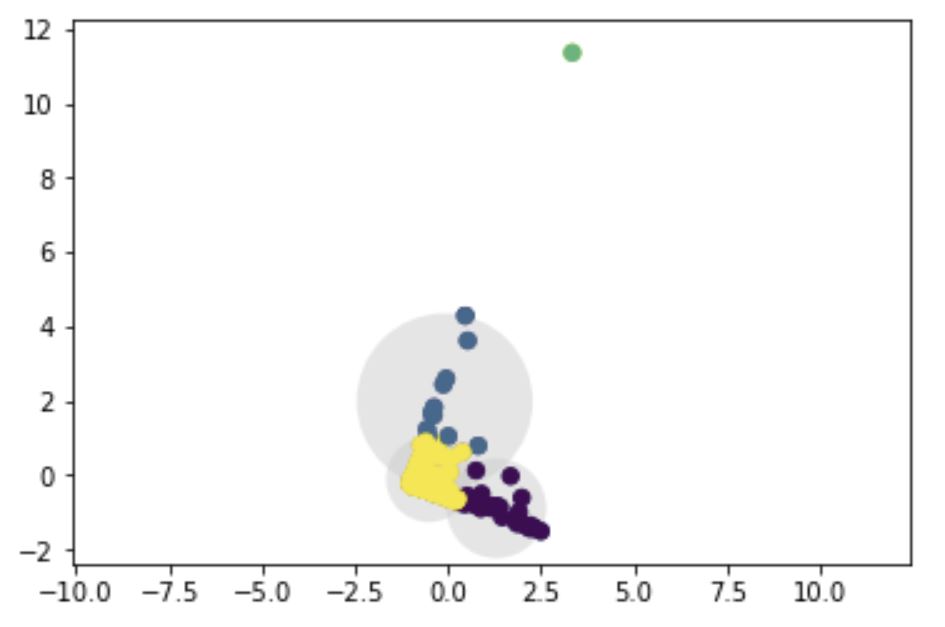
I am unclear about why k-means clustering can have overlap in clusters. From Chen (2018) I saw the following definition:
"..let the observations be a sample set to be partitioned into K disjoint clusters"
However I see an overlap in my plots, and am not sure why this is the case.
For reference, I am trying to cluster a multi-dimensional dataset with three variables (Recency, Frequency, Revenue). To visualize clustering, I can project 3D data into 2D using PCA and run k-means on that. Below is the code and plot I get:
df1=tx_user[["Recency","Frequency","Revenue"]]
#standardize
names = df1.columns
# Create the Scaler object
scaler = preprocessing.StandardScaler()
# Fit your data on the scaler object
scaled_df1 = scaler.fit_transform(df1)
df1 = pd.DataFrame(scaled_df1, columns=names)
df1.head()
del scaled_df1
sklearn_pca = PCA(n_components = 2)
X1 = sklearn_pca.fit_transform(df1)
X1 = X1[:, ::-1] # flip axes for better plotting
kmeans = KMeans(3, random_state=0)
labels = kmeans.fit(X1).predict(X1)
plt.scatter(X1[:, 0], X1[:, 1], c=labels, s=40, cmap='viridis');
from sklearn.cluster import KMeans
from scipy.spatial.distance import cdist
def plot_kmeans(kmeans, X, n_clusters=4, rseed=0, ax=None):
labels = kmeans.fit_predict(X)
# plot the input data
ax = ax or plt.gca()
ax.axis('equal')
#ax.set_ylim(-5000,7000)
ax.scatter(X[:, 0], X[:, 1], c=labels, s=40, cmap='viridis', zorder=2)
# plot the representation of the KMeans model
centers = kmeans.cluster_centers_
radii = [cdist(X[labels == i], [center]).max()
for i, center in enumerate(centers)]
for c, r in zip(centers, radii):
ax.add_patch(plt.Circle(c, r, fc='#CCCCCC', lw=3, alpha=0.5, zorder=1))
kmeans = KMeans(n_clusters=4, random_state=0)
plot_kmeans(kmeans, X1)
My question is: 1. Why is there an overlap? Is my clustering wrong if there is? 2. How does k-means decide cluster assignment incase there is an overlap?
Thank you
Reference: Chen, L., Xu, Z., Wang, H., & Liu, S. (2018). An ordered clustering algorithm based on K-means and the PROMETHEE method. International Journal of Machine Learning and Cybernetics, 9(6), 917-926.