I would like to create a map showing the bi-variate spatial correlation between two variables. This could be done either by doing a LISA map of bivariate Moran's I spatial correlation or using the L index proposed by Lee (2001).
The bi-variate Moran's I is not implemented in the spdep library, but the L index is, so here is what I've tried without success using the L index. A answer showing a solution based on Moran's I would also be very welcomed !
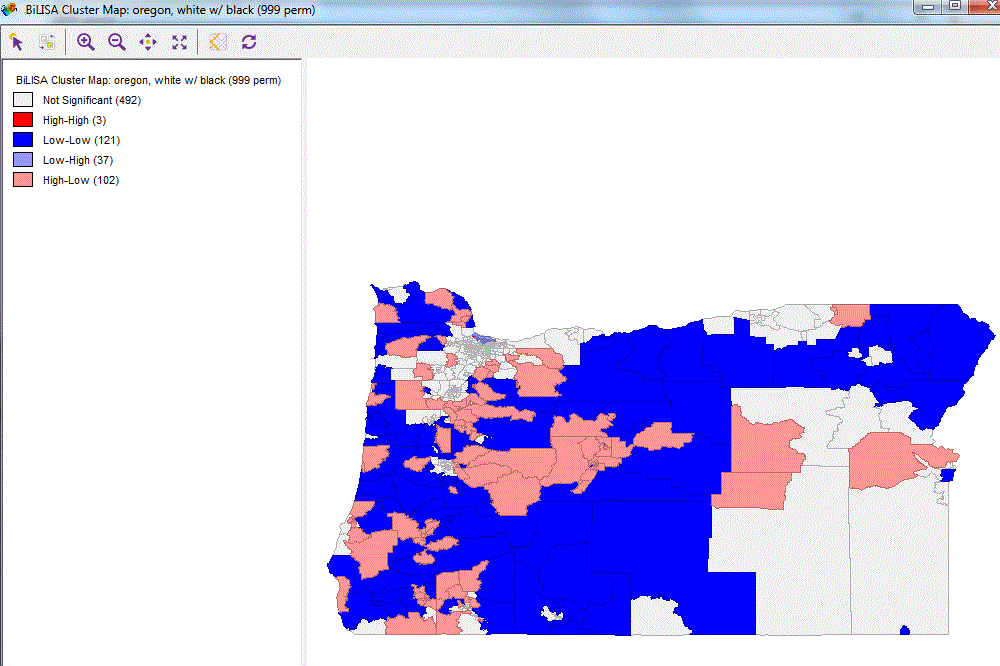
As you can see from the reproducible example below, I've manged so far to calculate the local L indexes. What I would like to do is to estimate the pseudo p-values and create a map of the results like those maps we use in LISA spatial clusters with high-high, high-low, ..., low-low.
In this example, the goal is to create a map with bi-variate Lisa association between black and white population. The map should be created in ggplot2 , showing the clusters:
- High-presence of black and High-presence of white people
- High-presence of black and Low-presence of white people
- Low-presence of black and High-presence of white people
- Low-presence of black and Low-presence oh white people
Reproducible example
library(UScensus2000tract)
library(ggplot2)
library(spdep)
library(sf)
# load data
data("oregon.tract")
# plot Census Tract map
plot(oregon.tract)
# Variables to use in the correlation: white and black population in each census track
x <- scale(oregon.tract$white)
y <- scale(oregon.tract$black)
# create Queen contiguity matrix and Spatial weights matrix
nb <- poly2nb(oregon.tract)
lw <- nb2listw(nb)
# Lee index
Lxy <-lee(x, y, lw, length(x), zero.policy=TRUE)
# Lee’s L statistic (Global)
Lxy[1]
#> -0.1865688811
# 10k permutations to estimate pseudo p-values
LMCxy <- lee.mc(x, y, nsim=10000, lw, zero.policy=TRUE, alternative="less")
# quik plot of local L
Lxy[[2]] %>% density() %>% plot() # Lee’s local L statistic (Local)
LMCxy[[7]] %>% density() %>% lines(col="red") # plot values simulated 10k times
# get confidence interval of 95% ( mean +- 2 standard deviations)
two_sd_above <- mean(LMCxy[[7]]) + 2 * sd(LMCxy[[7]])
two_sd_below <- mean(LMCxy[[7]]) - 2 * sd(LMCxy[[7]])
# convert spatial object to sf class for easier/faster use
oregon_sf <- st_as_sf(oregon.tract)
# add L index values to map object
oregon_sf$Lindex <- Lxy[[2]]
# identify significant local results
oregon_sf$sig <- if_else( oregon_sf$Lindex < 2*two_sd_below, 1, if_else( oregon_sf$Lindex > 2*two_sd_above, 1, 0))
# Map of Local L index but only the significant results
ggplot() + geom_sf(data=oregon_sf, aes(fill=ifelse( sig==T, Lindex, NA)), color=NA)



y_sthe spatially laggedy? – Autarch