I think you should reduce the number of %*% operators used in your calculation pipe, which should be the most straightforward and effective way to save time.
Code Optimization
For example, you can move your terms in the expression like below
exp(X %*% log(pr.t / (1 - pr.t)) + t(colSums(log(1 - pr.t)))[rep.int(1, nrow(X)), ]) %*% wts
where only the %*% operator is used only twice (instead of 3 as in your original scheme)
Benchmark
We compare your solution, @jblood94's solution, and my solution and see how the performance looks like
# pre-processing since they keep fixed during iterations
n <- ncol(X)
X0 <- unique(X)
mX0 <- 1 - X0
i <- match(X %*% 2^((n - 1):0), X0 %*% 2^((n - 1):0))
f_dhc <- function(X, pr.t, wts) {
mX <- 1 - X
exp(X %*% log(pr.t) + mX %*% log(1 - pr.t)) %*% wts
}
f_jblood94_1 <- function(X, pr.t, wts) {
(exp(X0 %*% log(pr.t) + mX0 %*% log1p(-pr.t)) %*% wts)[i, , drop = FALSE]
}
f_jblood94_2 <- function(X, pr.t, wts) {
(exp(X0 %*% log(pr.t / (1 - pr.t)) + t(colSums(log1p(-pr.t)))[rep.int(1, nrow(X0)), ]) %*% wts)[i, , drop = FALSE]
}
f_TIC1 <- function(X, pr.t, wts) {
exp(X %*% log(pr.t / (1 - pr.t)) + t(colSums(log(1 - pr.t)))[rep.int(1, nrow(X)), ]) %*% wts
}
f_TIC2 <- function(X, pr.t, wts) {
exp(X %*% log(pr.t / (1 - pr.t)) + outer(rep.int(1, nrow(X)), colSums(log(1 - pr.t)))) %*% wts
}
f_TIC3 <- function(X, pr.t, wts) {
exp(X %*% log(pr.t / (1 - pr.t)) + kronecker(matrix(rep.int(1, nrow(X))), t(colSums(log(1 - pr.t))))) %*% wts
}
f_TIC4 <- function(X, pr.t, wts) {
exp(X %*% qlogis(pr.t) + t(colSums(log1p(-pr.t)))[rep.int(1, nrow(X)), ]) %*% wts
}
mbm <- microbenchmark(
dhc = f_dhc(X, pr.t, wts),
jblood94_1 = f_jblood94_1(X, pr.t, wts),
jblood94_2 = f_jblood94_2(X, pr.t, wts),
TIC1 = f_TIC1(X, pr.t, wts),
TIC2 = f_TIC2(X, pr.t, wts),
TIC3 = f_TIC3(X, pr.t, wts),
TIC4 = f_TIC4(X, pr.t, wts),
check = "equal",
unit = "relative"
)
set.seed(0)
p <- 1000
q <- 5
r <- 15
X <- matrix(sample(c(0, 1), p * q, replace = TRUE), p, q)
pr.t <- matrix(runif(r * q), q, r)
wts <- runif(r)
and we will see
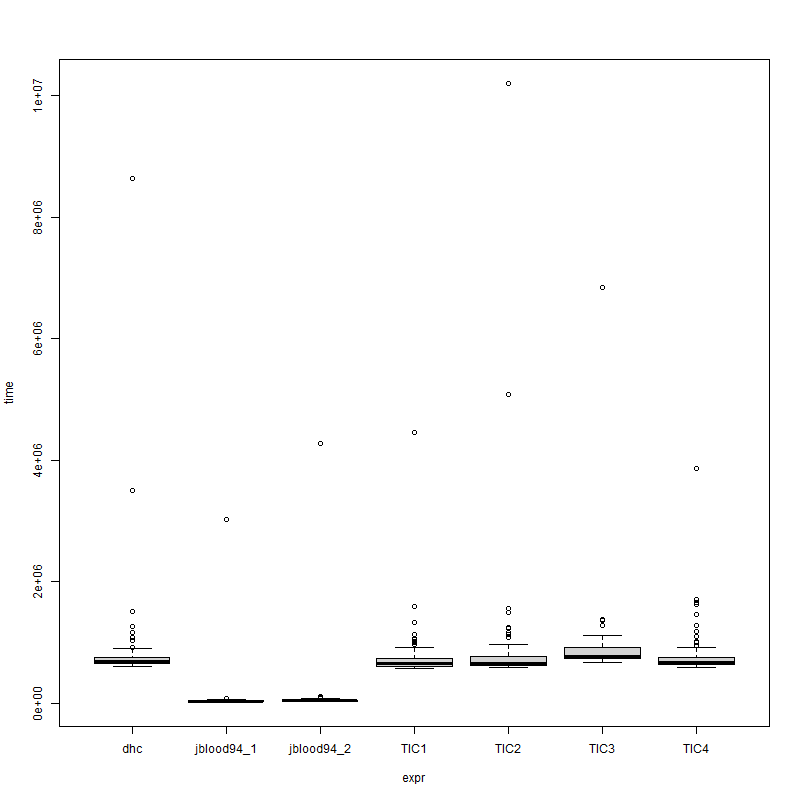
> mbm
Unit: relative
expr min lq mean median uq max neval
dhc 22.869560 22.049715 19.49625 19.354935 17.437032 20.113939 100
jblood94_1 1.000000 1.000000 1.00000 1.000000 1.000000 1.000000 100
jblood94_2 1.255509 1.448805 1.59287 1.521874 1.652512 1.655629 100
TIC1 22.380653 20.909609 10.14739 18.157829 17.205723 1.243090 100
TIC2 22.621384 21.141098 11.76505 18.266085 17.322903 3.590901 100
TIC3 26.265694 25.490583 12.36498 22.410242 19.683481 1.743739 100
TIC4 22.921410 21.263929 10.45764 19.084340 17.894608 1.141133 100
and plot(mbm) shows
![enter image description here]()
set.seed(0)
p <- 1000
q <- 1000
r <- 100
X <- matrix(sample(c(0, 1), p * q, replace = TRUE), p, q)
pr.t <- matrix(runif(r * q), q, r)
wts <- runif(r)
and we will see
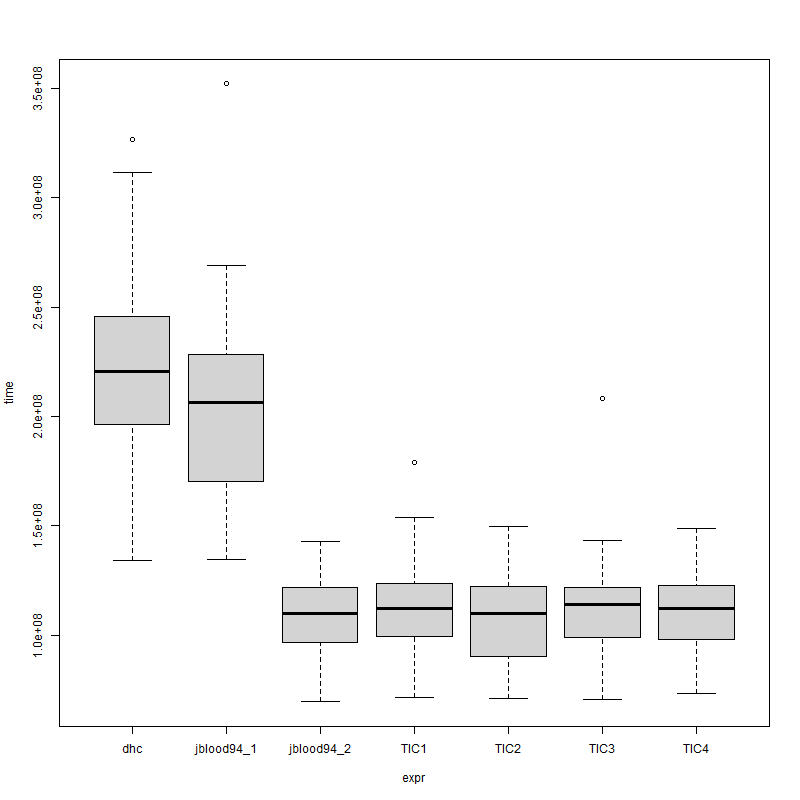
> mbm
Unit: relative
expr min lq mean median uq max neval
dhc 1.8862821 2.172686 2.030361 2.005938 2.0100325 2.1832300 100
jblood94_1 1.8933325 1.884341 1.881695 1.875507 1.8684887 2.3552021 100
jblood94_2 0.9776827 1.071931 1.016522 1.000148 0.9974477 0.9546140 100
TIC1 1.0086822 1.099217 1.035331 1.018304 1.0093273 1.1968782 100
TIC2 1.0000000 1.000000 1.000000 1.000000 1.0000000 1.0000000 100
TIC3 0.9939343 1.095266 1.038841 1.037027 0.9967151 1.3936333 100
TIC4 1.0297307 1.084197 1.024587 1.018200 1.0050159 0.9950423 100
and plot(mbm) shows
![enter image description here]()

XandmXalways binary matrices? – DalesmanMatrix: it doesn't seem to help in this case, even when I increase sparsity to 99%. As @Dalesman hints, an indexing-based solution might help? – Rampage