I am attempting to implement a CNN-LSTM that classifies mel-spectrogram images representing the speech of people with Parkinson's Disease/Healthy Controls. I am trying to implement a pre-existing model (DenseNet-169) with an LSTM model, however I am running into the following error: ValueError: Input 0 of layer zero_padding2d is incompatible with the layer: expected ndim=4, found ndim=3. Full shape received: [None, 216, 1]. Can anyone advise where I'm going wrong?
import librosa
import os
import glob
import IPython.display as ipd
from pathlib import Path
import timeit
import time, sys
%matplotlib inline
import matplotlib.pyplot as plt
import librosa.display
import pandas as pd
from sklearn import datasets, linear_model
from sklearn.model_selection import train_test_split
from matplotlib import pyplot as plt
import numpy as np
import cv2
import seaborn as sns
%tensorflow_version 1.x #version 1 works without problems
import tensorflow
from tensorflow.keras import models
from sklearn.preprocessing import LabelEncoder
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.layers import TimeDistributed
import keras
from tensorflow.keras.models import Sequential
from tensorflow.keras.callbacks import EarlyStopping
from sklearn.metrics import confusion_matrix, plot_confusion_matrix
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dropout, Dense, BatchNormalization, Activation, GaussianNoise, LSTM
from sklearn.metrics import accuracy_score
DATA_DIR = Path('/content/drive/MyDrive/PhD_Project_Experiments/Spontaneous_Dialogue_PD_Dataset')
diagnosis = [x.name for x in DATA_DIR.glob('*') if x.is_dir()]
diagnosis
def create_paths_ds(paths: Path, label: str) -> list:
EXTENSION_TYPE = '.wav'
return [(x, label) for x in paths.glob('*' + EXTENSION_TYPE)]
from collections import Counter
categories_to_use = [
'Parkinsons_Disease',
'Healthy_Control',
]
NUM_CLASSES = len(categories_to_use)
print(f'Number of classes: {NUM_CLASSES}')
paths_all_labels = []
for cat in categories_to_use:
paths_all_labels += create_paths_ds(DATA_DIR / cat, cat)
X_train, X_test = train_test_split(paths_all_labels,test_size=0.1, stratify = [paths_all_labels[y][1] for y in range(len(paths_all_labels))] ) #fix stratified sampling for test data
X_train, X_val = train_test_split(X_train, test_size=0.2, stratify = [X_train[y][1] for y in range(len(X_train))] )
for i in categories_to_use:
print('Number of train samples for '+i+': '+ str([X_train[y][1] for y in range(len(X_train))].count(i))) #checks whether train samples are equally divided
print('Number of test samples for '+i+': '+ str([X_test[y][1] for y in range(len(X_test))].count(i))) #checks whether test samples are equally divided
print('Number of validation samples for '+i+': '+ str([X_val[y][1] for y in range(len(X_val))].count(i))) #checks whether val samples are equally divided
print(f'Train length: {len(X_train)}')
print(f'Validation length: {len(X_val)}')
print(f'Test length: {len(X_test)}')
def load_and_preprocess_lstm(dataset, SAMPLE_SIZE = 30):
IMG_SIZE = (216,128)
progress=0
data = []
labels = []
for (path, label) in dataset:
audio, sr = librosa.load(path)
dur = librosa.get_duration(audio, sr = sr)
sampleNum = int(dur / SAMPLE_SIZE)
offset = (dur % SAMPLE_SIZE) / 2
for i in range(sampleNum):
audio, sr = librosa.load(path, offset= offset+i, duration=SAMPLE_SIZE)
sample = librosa.feature.melspectrogram(audio, sr=sr)
# print(sample.shape)
sample = cv2.resize(sample, dsize=IMG_SIZE)
sample = np.expand_dims(sample,-1)
print(sample.shape)
data += [(sample, label)]
labels += [label]
progress +=1
print('\r Progress: '+str(round(100*progress/len(dataset))) + '%', end='')
return data, labels
def retrieve_samples(sample_size, model_type):
if model_type == 'cnn':
print("\nLoading train samples")
X_train_samples, train_labels = load_and_preprocess_cnn(X_train,sample_size)
print("\nLoading test samples")
X_test_samples, test_labels = load_and_preprocess_cnn(X_test,sample_size)
print("\nLoading val samples")
X_val_samples, val_labels = load_and_preprocess_cnn(X_val,sample_size)
print('\n')
elif model_type == 'lstm':
print("\nLoading train samples")
X_train_samples, train_labels = load_and_preprocess_lstm(X_train,sample_size)
print("\nLoading test samples")
X_test_samples, test_labels = load_and_preprocess_lstm(X_test,sample_size)
print("\nLoading val samples")
X_val_samples, val_labels = load_and_preprocess_lstm(X_val,sample_size)
print('\n')
elif model_type == "cnnlstm":
print("\nLoading train samples")
X_train_samples, train_labels = load_and_preprocess_lstm(X_train,sample_size)
print("\nLoading test samples")
X_test_samples, test_labels = load_and_preprocess_lstm(X_test,sample_size)
print("\nLoading val samples")
X_val_samples, val_labels = load_and_preprocess_lstm(X_val,sample_size)
print('\n')
print("shape: " + str(X_train_samples[0][0].shape))
print("number of training samples: "+ str(len(X_train_samples)))
print("number of validation samples: "+ str(len(X_val_samples)))
print("number of test samples: "+ str(len(X_test_samples)))
return X_train_samples, X_test_samples, X_val_samples
def create_cnn_lstm_model(input_shape):
model = Sequential()
cnn = tensorflow.keras.applications.DenseNet169(include_top=True, weights=None, input_tensor=None, input_shape=input_shape, pooling=None, classes=2)
# define LSTM model
model.add(tensorflow.keras.layers.TimeDistributed(cnn, input_shape=input_shape))
model.add(LSTM(units = 512, dropout=0.5, recurrent_dropout=0.3, return_sequences = True, input_shape = input_shape))
model.add(LSTM(units = 512, dropout=0.5, recurrent_dropout=0.3, return_sequences = False))
model.add(Dense(units=NUM_CLASSES, activation='sigmoid'))#Compile
model.compile(loss=tensorflow.keras.losses.binary_crossentropy, optimizer='adam', metrics=['accuracy'])
print(model.summary())
return model
def create_model_data_and_labels(X_train_samples, X_val_samples, X_test_samples):
#Prepare samples to work for training the model
labelizer = LabelEncoder()
#prepare training data and labels
x_train = np.array([x[0] for x in X_train_samples])
y_train = np.array([x[1] for x in X_train_samples])
y_train = labelizer.fit_transform(y_train)
y_train = to_categorical(y_train)
#prepare validation data and labels
x_val = np.array([x[0] for x in X_val_samples])
y_val = np.array([x[1] for x in X_val_samples])
y_val = labelizer.transform(y_val)
y_val = to_categorical(y_val)
#prepare test data and labels
x_test = np.array([x[0] for x in X_test_samples])
y_test = np.array([x[1] for x in X_test_samples])
y_test = labelizer.transform(y_test)
y_test = to_categorical(y_test)
return x_train, y_train, x_val, y_val, x_test, y_test, labelizer
#Main loop for testing multiple sample sizes
#choose model type: 'cnn' or 'lstm'
model_type = 'cnnlstm'
n_epochs = 20
patience= 20
es = EarlyStopping(patience=20)
fragment_sizes = [5,10]
start = timeit.default_timer()
ModelData = pd.DataFrame(columns = ['Model Type','Fragment size (s)', 'Time to Compute (s)', 'Early Stopping epoch', 'Training accuracy', 'Validation accuracy', 'Test Accuracy']) #create a DataFrame for storing the results
conf_matrix_data = []
for i in fragment_sizes:
start_per_size = timeit.default_timer()
print(f'\n---------- Model trained on fragments of size: {i} seconds ----------------')
X_train_samples, X_test_samples, X_val_samples = retrieve_samples(i,model_type)
x_train, y_train, x_val, y_val, x_test, y_test, labelizer = create_model_data_and_labels(X_train_samples, X_val_samples, X_test_samples)
if model_type == 'cnn':
model = create_cnn_model(X_train_samples[0][0].shape)
elif model_type == 'lstm':
model = create_lstm_model(X_train_samples[0][0].shape)
elif model_type == 'cnnlstm':
model = create_cnn_lstm_model(X_train_samples[0][0].shape)
history = model.fit(x_train, y_train,
batch_size = 8,
epochs=n_epochs,
verbose=1,
callbacks=[es],
validation_data=(x_val, y_val))
print('Finished training')
early_stopping_epoch = len(history.history['accuracy'])
training_accuracy = history.history['accuracy'][early_stopping_epoch-1-patience]
validation_accuracy = history.history['val_accuracy'][early_stopping_epoch-1-patience]
plot_data(history, i)
predictions = model.predict(x_test)
score = accuracy_score(labelizer.inverse_transform(y_test.argmax(axis=1)), labelizer.inverse_transform(predictions.argmax(axis=1)))
print('Fragment size = ' + str(i) + ' seconds')
print('Accuracy on test samples: ' + str(score))
conf_matrix_data += [(predictions, y_test, i)]
stop_per_size = timeit.default_timer()
time_to_compute = round(stop_per_size - start_per_size)
print ('Time to compute: '+str(time_to_compute))
ModelData.loc[len(ModelData)] = [model_type, i, time_to_compute, early_stopping_epoch, training_accuracy, validation_accuracy, score] #store particular settings configuration, early stoppping epoch and accuracies in dataframe
stop = timeit.default_timer()
print ('\ntime to compute: '+str(stop-start))

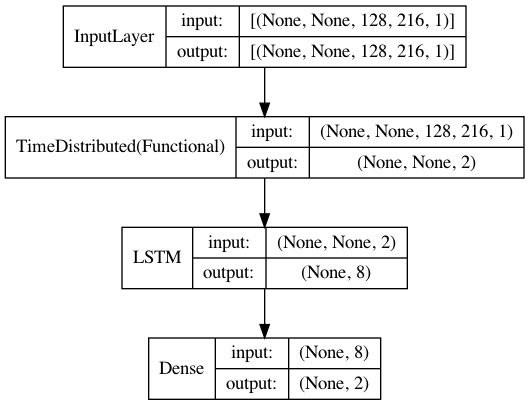
create_cnn_lstm_model()function. What is yourinput_shapethat you use as parameter to call the funciton – KerianneX_train_samples[0][0].shapeand tell me? – Keriannecnnlayer. In this step -tensorflow.keras.layers.TimeDistributed(cnn, input_shape=(128,216,1))), you are passing the 128 dimension axis as time axis. That means each of the cnn is left with(216,1), which is not an image and therefore throws an error because its expecting a 3D tensors and not 2D. – Kerianne(128, 216, 1)becomes an image input for the cnn. – Kerianne