I have an input csv file with 4500 rows. Each row has a unique ID and for each row, I have to read some data, do some calculation, and write the output in a csv file so that I have 4500 csv files written in my output directory. An individual output csv file contains a single row of data with 8 columns
Since I have to perform the same calculation on each row of my input csv, I thought I can parallelise this task using foreach. Following is the overall structure of the logic
library(doSNOW)
library(foreach)
library(data.table)
input_csv <- fread('inputFile.csv'))
# to track the progres of the loop
iterations <- nrow(input_csv)
pb <- txtProgressBar(max = iterations, style = 3)
progress <- function(n) setTxtProgressBar(pb, n)
opts <- list(progress = progress)
myClusters <- makeCluster(6)
registerDoSNOW(myClusters)
results <-
foreach(i = 1:nrow(input_csv),
.packages = c("myCustomPkg","dplyr","arrow","zoo","data.table","rlist","stringr"),
.errorhandling = 'remove',
.options.snow = opts) %dopar%
{
rowRef <- input_csv[i, ]
# read data for the unique location in `rowRef`
weather.path <- arrow(paste0(rowRef$locationID'_weather.parquet')))
# do some calculations
# save the results as csv
fwrite(temp_result, file.path(paste0('output_iter_',i,'.csv')))
return(temp_result)
}
The above code works fine but always get stuck/inactive/does not do anything after finishing 25% or 30% of the rows in input_csv. I keep looking at my output directory that after N% of iterations, no file is being written. I suspect if the foreach loop goes into some sleep mode? What I find more confounding is that if I kill the job, re-run the above code, it does say 16% or 30% and then goes inactive again i.e. with each fresh run, it "sleeps" at different progress level.
I can't figure out how to give a minimal reproducible example in this case but thought if anyone knows of any checklist I should go through or potential issues that is causing this would be really helpful. Thanks
EDIT I am still struggling with this issue. If there is any more information I can provide, please let me know.
EDIT2
My original inputFile contains 213164 rows. So I split my the big file
into 46 smaller files so that each file has 4634 rows
library(foreach)
library(data.table)
library(doParallel)
myLs <- split(mydat, (as.numeric(rownames(mydat))-1) %/% 46))
Then I did this:
for(pr in 1:46){
input_csv <- myLs[[pr]]
myClusters <- parallel::makeCluster(6)
doParallel::registerDoParallel(myClusters)
results <-
foreach(i = 1:nrow(input_csv),
.packages = c("myCustomPkg","dplyr","arrow","zoo","data.table","rlist","stringr"),
.errorhandling = 'remove',
.verbose = TRUE) %dopar%
{
rowRef <- input_csv[i, ]
# read data for the unique location in `rowRef`
weather.path <- arrow(paste0(rowRef$locationID'_weather.parquet')))
# do some calculations
# save the results as csv
fwrite(temp_result, file.path(paste0('output_iter_',i,'_',pr,'.csv')))
gc()
}
parallel::stopCluster(myClusters)
gc()
}
This too works till say pr = 7 or pr = 8 iteration and then does not proceed and also does not generate any error message. I am so confused.
EDIT this is what my CPU usage looks like. I only used 4 cores to generate this image. Will anyone be able to explain if there's anything in this image that might address my question.

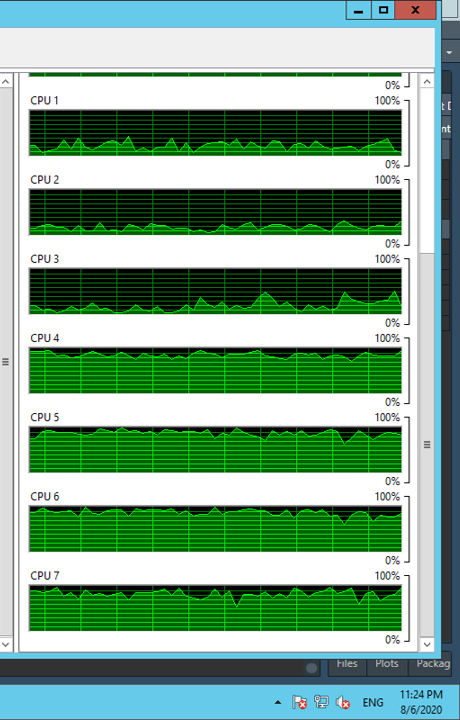
temp_result. Is it a memory issue? – Courageousfwrite(), but it looks like they deleted the question. If I remember correctly, it was faster for e.g., 50 files but slower for e.g., 500 files. I cannot rememebr the magnitude of the difference. All that to say, it may be worth trying to swap outfwrite()forreadr::write_csv(). One other possibility, is that you can try to write the files in another step considering you save them all toresults– Moritareadrbut it also did not work. I have also edited my question slightly now to give some more information – Intitulegc(), tryrm(temp_result). Did you monitor the memory usage for your edited code? You don't need to do this in R, you can also use the system tools. I've also had it before that the garbage collector didn't really work for parallel processing, so it can be a bit tricky – DextroamphetaminesessionInfo()and environment variables for the parent environment and a parallel task? My sense is you want to try running your code outside of RStudio. Why? If the code is run inside RStudio, your system calls will pass through RStudio and be part of its memory footpad. Secondly, remember that you load the packages in each parallel task. Is your packagemyCustomPkgparallel safe? – Bight# do some calculationsand# save the results as csv. – Bight