It is not RGB pixel array and the better way is converting to gray image.
The way to get CT Image is to get the attribute of pixel_array in CT dicom file.
The type of elements in pixel_array of CT dicom file are all uint16.But a lot of tool in python, like OpenCV, Some AI stuff, cannot be compatible with the type.
After getting pixel_array (CT Image) from CT dicom file, you always need to convert the pixel_array into gray image, so that you can process this gray image by a lot of image processing tool in python.
The following code is a working example to convert pixel_array into gray image.
import matplotlib.pyplot as plt
import os
import pydicom
import numpy as np
# Abvoe code is to import dependent libraries of this code
# Read some CT dicom file here by pydicom library
ct_filepath = r"<YOUR_CT_DICOM_FILEPATH>"
ct_dicom = pydicom.read_file(ct_filepath)
img = ct_dicom.pixel_array
# Now, img is pixel_array. it is input of our demo code
# Convert pixel_array (img) to -> gray image (img_2d_scaled)
## Step 1. Convert to float to avoid overflow or underflow losses.
img_2d = img.astype(float)
## Step 2. Rescaling grey scale between 0-255
img_2d_scaled = (np.maximum(img_2d,0) / img_2d.max()) * 255.0
## Step 3. Convert to uint
img_2d_scaled = np.uint8(img_2d_scaled)
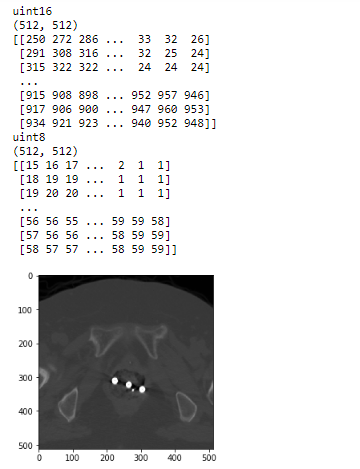
# Show information of input and output in above code
## (1) Show information of original CT image
print(img.dtype)
print(img.shape)
print(img)
## (2) Show information of gray image of it
print(img_2d_scaled.dtype)
print(img_2d_scaled.shape)
print(img_2d_scaled)
## (3) Show the scaled gray image by matplotlib
plt.imshow(img_2d_scaled, cmap='gray', vmin=0, vmax=255)
plt.show()
And the following is result of what I print out.
![enter image description here]()

pixel_array.shape? – KempchanR = pixel_array[:,:,0]. – KempPhotometric Interpretation, that does not mean order. – Neurath