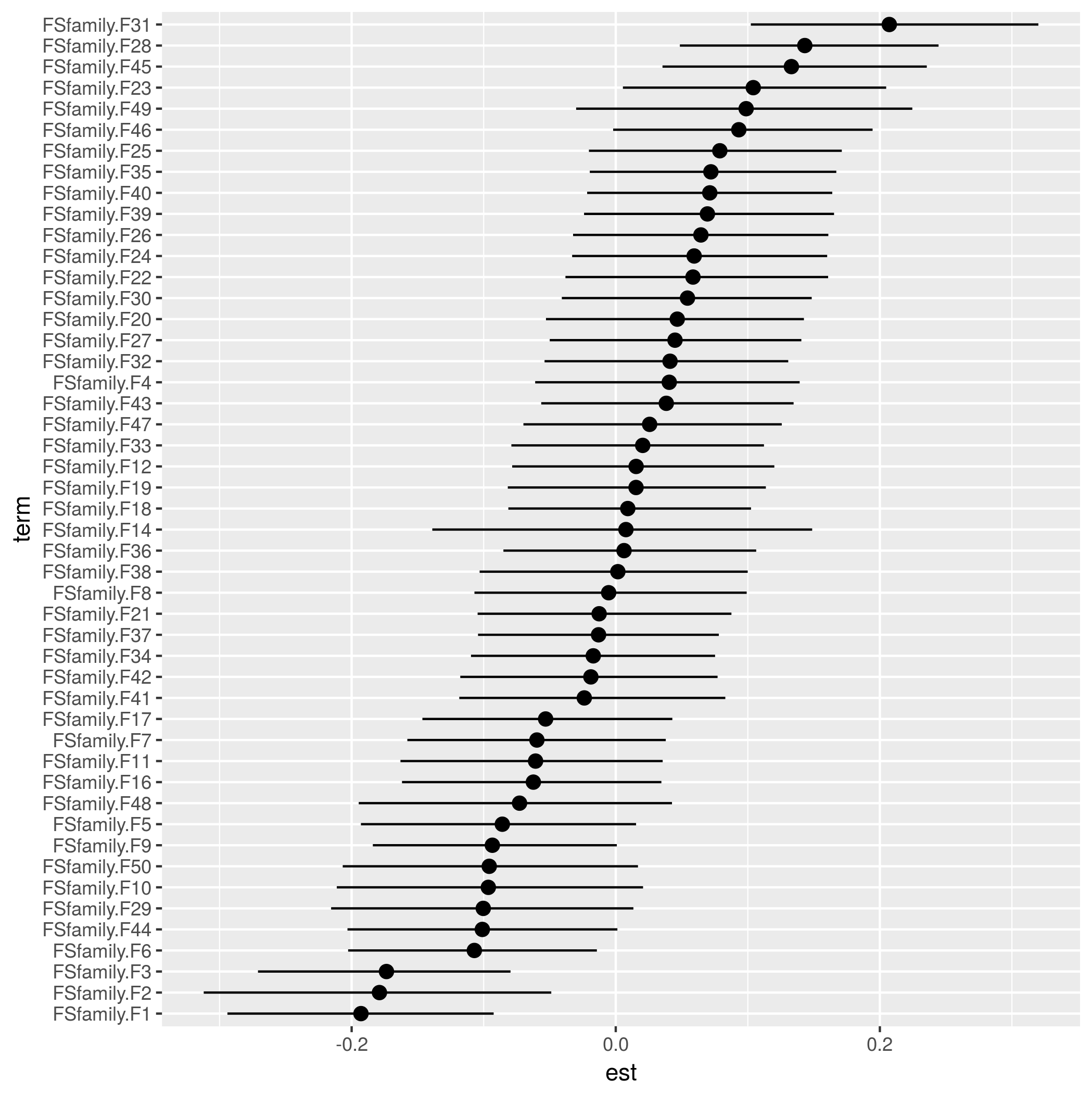
I am looking for a command similar to ranef() used in nlme, lme4, and brms that will allow me to extract the individual random effects in my MCMCglmm model. In my dataset, I have 40 providers and I would like to extract the random effects for each provider and plot them in a caterpillar plot. Any suggestions would be great. Thank you.
In case it is helpful, here is my MCMCglmm model:
prior.3 <- list(R = list(R1 = list(V = diag(2), nu = 0.002)),
G = list(G1 = list(V = diag(2), nu = 0.002),
G2 = list(V = diag(2), nu = 0.002)))
mc_mod2 <- MCMCglmm(outcome ~ 1, data = filter(data, rem2 == "white" | rem2 == "rem"),
random = ~ idh(rem2):id + us(rem2):provider,
rcov = ~idh(rem2):units,
verbose = TRUE,
prior = prior.3,
family = "gaussian",
nitt = 100000, burnin = 5000,
pr = TRUE)

ranef(mc_mod2)and I am thrown the following error:Error in UseMethod("ranef") : no applicable method for 'ranef' applied to an object of class "MCMCglmm"– Frodeen