According to SimpleITK's documentation, the process of image resampling involves 4 steps:
- Image - the image we resample, given in the coordinate system;
- Resampling grid - a regular grid of points given in a coordinate system which will be mapped to the coordinate system;
- Transformation - maps points from the coordinate system to coordinate system;
- Interpolator - a method for obtaining the intensity values at arbitrary points in the coordinate system from the values of the points defined by the Image
The following snippet is for downsampling the image preserving its coordinate system properties:
def downsamplePatient(patient_CT, resize_factor):
original_CT = sitk.ReadImage(patient_CT,sitk.sitkInt32)
dimension = original_CT.GetDimension()
reference_physical_size = np.zeros(original_CT.GetDimension())
reference_physical_size[:] = [(sz-1)*spc if sz*spc>mx else mx for sz,spc,mx in zip(original_CT.GetSize(), original_CT.GetSpacing(), reference_physical_size)]
reference_origin = original_CT.GetOrigin()
reference_direction = original_CT.GetDirection()
reference_size = [round(sz/resize_factor) for sz in original_CT.GetSize()]
reference_spacing = [ phys_sz/(sz-1) for sz,phys_sz in zip(reference_size, reference_physical_size) ]
reference_image = sitk.Image(reference_size, original_CT.GetPixelIDValue())
reference_image.SetOrigin(reference_origin)
reference_image.SetSpacing(reference_spacing)
reference_image.SetDirection(reference_direction)
reference_center = np.array(reference_image.TransformContinuousIndexToPhysicalPoint(np.array(reference_image.GetSize())/2.0))
transform = sitk.AffineTransform(dimension)
transform.SetMatrix(original_CT.GetDirection())
transform.SetTranslation(np.array(original_CT.GetOrigin()) - reference_origin)
centering_transform = sitk.TranslationTransform(dimension)
img_center = np.array(original_CT.TransformContinuousIndexToPhysicalPoint(np.array(original_CT.GetSize())/2.0))
centering_transform.SetOffset(np.array(transform.GetInverse().TransformPoint(img_center) - reference_center))
centered_transform = sitk.Transform(transform)
centered_transform.AddTransform(centering_transform)
# sitk.Show(sitk.Resample(original_CT, reference_image, centered_transform, sitk.sitkLinear, 0.0))
return sitk.Resample(original_CT, reference_image, centered_transform, sitk.sitkLinear, 0.0)
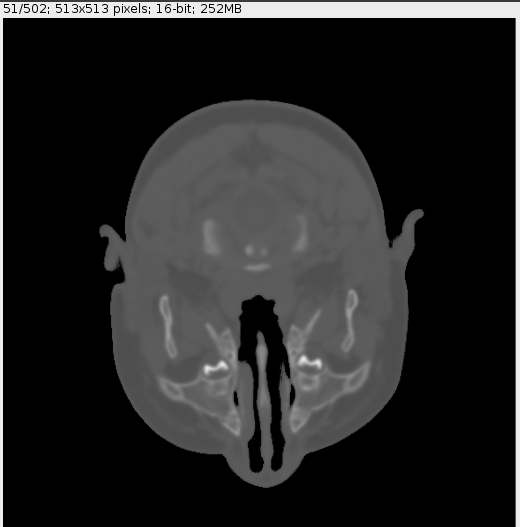
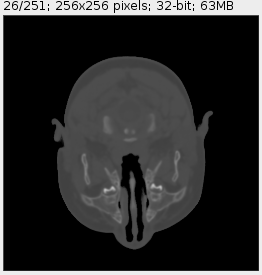
Using the snippet above in a brain CT scan we get:
![Original CT scan]()
![Downsampled CT scan]()