I want to compare two nested linear models, call them m01, and m02 where m01 is the reduced model and m02 is the full model. I want to do a simple F-test to see if the full model adds significant utility over the reduced model.
This is very simple in R. For example:
mtcars <- read.csv("https://raw.githubusercontent.com/focods/WonderfulML/master/data/mtcars.csv")
m01 <- lm(mpg ~ am + wt, mtcars)
m02 <- lm(mpg ~ am + am:wt, mtcars)
anova(m01, m02)
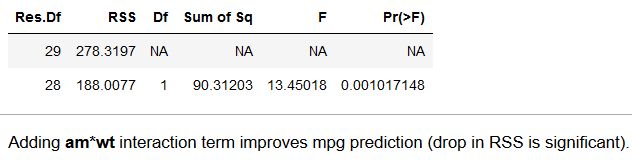
Gives me the following output:
Which tells me that adding the am: wt interaction term significantly improves the model. Is there a way to do something similar to this in Python/sklearn/statsmodels?
Edit: I looked at this question before posting this one and can not figure out how they are the same. The other question is doing an F-test on two vectors. This question is about comparing 2 nested linear models.
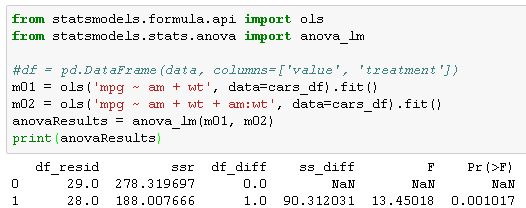
I think this is what I need:
but am not sure what exactly to pass this function. If anyone could provide or point to an example, that would be extremely helpful.