I'm switching from Mathematica to R but I'm finding some difficulties with visualizations.
I'm trying to do a heatmap as follows:
short
penetration scc pi0
1 0 0 0.002545268
2 5 0 -0.408621176
3 10 0 -0.929432006
4 15 0 -1.121309680
5 20 0 -1.587298317
6 25 0 -2.957853131
7 30 0 -5.123329738
8 0 50 1.199748327
9 5 50 0.788581883
10 10 50 0.267771053
11 15 50 0.075893379
12 20 50 -0.390095258
13 25 50 -1.760650073
14 30 50 -3.926126679
15 0 100 2.396951386
16 5 100 1.985784941
17 10 100 1.464974112
18 15 100 1.273096438
19 20 100 0.807107801
20 25 100 -0.563447014
21 30 100 -2.728923621
mycol <- c("navy", "blue", "cyan", "lightcyan", "yellow", "red", "red4")
ggplot(data = short, aes(x = penetration, y = scc)) +
geom_tile(aes(fill = pi0)) +
scale_fill_gradientn(colours = mycol)
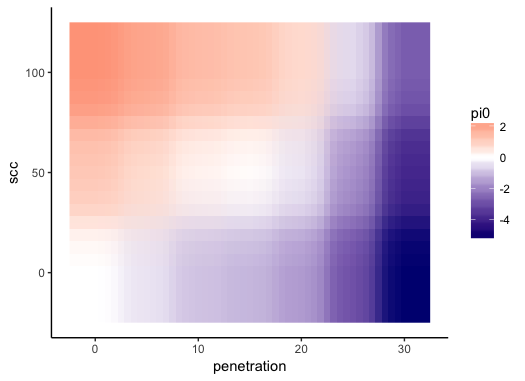
And I get this:

But I need something like this:

That is, I would like that the color is continuous (degraded) over the surface of the plot instead of discrete for each square. I've seen in other SO questions that some people interpolate de data but I think there should be an easier way to do it inside the ggplot call (in Mathematica is done by default).
Besides, I would like to lock the color scale such that the 0 is always white (separating therefore between warm colors for positive values and cold for negative ones) and the color distribution is always the same across plots independently of the range of the data (since I will use the same plot structure for several datasets)

scale_fill_gradient2(midpoint = 0, low = "your negative color", mid = "white", high = "your positive color"). That constrains zero to always be white. – Soliloquy