I'm tyring to make a graph in which different link categories have different colors, I use this as input (sample):
source target cat weight
<fctr> <fctr> <chr> <dbl>
1 human water 1 30
2 human water 2 49
3 human water 3 2
4 human water 4 14
5 human water 5 19
Then I transform this data into nodes, links and link.colors and plot it, this will give:
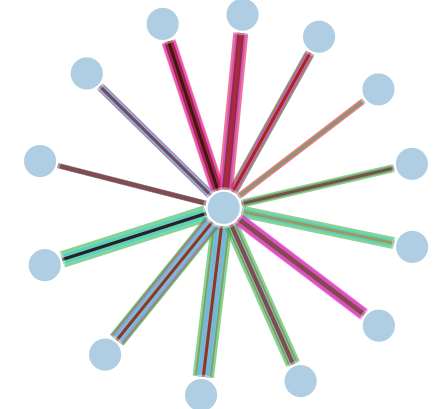
As you can see in this network, the links are plotted on top of each other. Therefore, it's impossible to see the differences. So I want to plot the links next to each other instead of on top of each other.
I can't provide the whole dataset, but a simple example will but sufficient I suppose:
library(networkD3)
link <- read.table(text = ' source target value
1 0 1 30
2 0 1 49', header = T)
nodes <- read.table(text = ' name group size
1 human 1 10
2 water 1 10', header = T)
link.colors <- c('#e6194b','#3cb44b')
network <- forceNetwork(Links = link, Nodes = nodes,
Source = "source", Target = "target",
Value = "value", NodeID = "name",
Group = "group", opacity = 0.6,
zoom = TRUE, charge = -60,
fontSize = 15, linkColour = link.colors)
# plot in R
network
# zooming doesn't work in Rstudio for me so I write it to a file instead of plotting it
# and then open it in my browser
saveNetwork(network, file = 'network_example.html', selfcontained = FALSE)
This code will produce the left network, whereas I want to have something like the right one
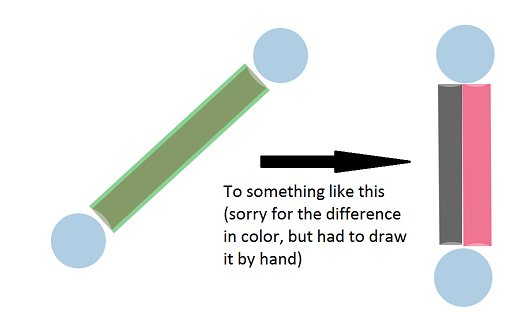
not sure if this is R or more JS, therefore I tagged it as both

saveNetwork()workaround,... – Surveying