Main issue: Why coefficients of Lasso regression are not shrunk to zero with minimization done by scipy.minimize?
I am trying to create Lasso model, using scipy.minimize. However, it is working only when alpha is zero (thus only like basic squared error). When alpha is not zero, it returns worse result (higher loss) and still none of coefficients is zero.
I know that Lasso is not differentiable, but I tried to use Powell optimizer, that should handle non-differential loss (also I tried BFGS, that should handle non-smooth). None of these optimizers worked.
For testing this, I created dataset where y is random (provided here to be reproducible), first feature of X is exactly y*.5 and other four features are random (also provided here to be reproducible). I would expect the algorithm to shrink these random coefficients to zero and keep only the first one, but it's not happening.
For lasso loss function I am using formula from this paper (figure 1, first page)
My code is following:
from scipy.optimize import minimize
import numpy as np
class Lasso:
def _pred(self,X,w):
return np.dot(X,w)
def LossLasso(self,weights,X,y,alpha):
w = weights
yp = self._pred(X,w)
loss = np.linalg.norm(y - yp)**2 + alpha * np.sum(abs(w))
return loss
def fit(self,X,y,alpha=0.0):
initw = np.random.rand(X.shape[1]) #initial weights
res = minimize(self.LossLasso,
initw,
args=(X,y,alpha),
method='Powell')
return res
if __name__=='__main__':
y = np.array([1., 0., 1., 0., 0., 1., 1., 0., 0., 0., 1., 0., 0., 0., 1., 0., 1.,
1., 1., 0.])
X_informative = y.reshape(20,1)*.5
X_noninformative = np.array([[0.94741352, 0.892991 , 0.29387455, 0.30517762],
[0.22743465, 0.66042825, 0.2231239 , 0.16946974],
[0.21918747, 0.94606854, 0.1050368 , 0.13710866],
[0.5236064 , 0.55479259, 0.47711427, 0.59215551],
[0.07061579, 0.80542011, 0.87565747, 0.193524 ],
[0.25345866, 0.78401146, 0.40316495, 0.78759134],
[0.85351906, 0.39682136, 0.74959904, 0.71950502],
[0.383305 , 0.32597392, 0.05472551, 0.16073454],
[0.1151415 , 0.71683239, 0.69560523, 0.89810466],
[0.48769347, 0.58225877, 0.31199272, 0.37562258],
[0.99447288, 0.14605177, 0.61914979, 0.85600544],
[0.78071238, 0.63040498, 0.79964659, 0.97343972],
[0.39570225, 0.15668933, 0.65247826, 0.78343458],
[0.49527699, 0.35968554, 0.6281051 , 0.35479879],
[0.13036737, 0.66529989, 0.38607805, 0.0124732 ],
[0.04186019, 0.13181696, 0.10475994, 0.06046115],
[0.50747742, 0.5022839 , 0.37147486, 0.21679859],
[0.93715221, 0.36066077, 0.72510501, 0.48292022],
[0.47952644, 0.40818585, 0.89012395, 0.20286356],
[0.30201193, 0.07573086, 0.3152038 , 0.49004217]])
X = np.concatenate([X_informative,X_noninformative],axis=1)
#alpha zero
clf = Lasso()
print(clf.fit(X,y,alpha=0.0))
#alpha nonzero
clf = Lasso()
print(clf.fit(X,y,alpha=0.5))
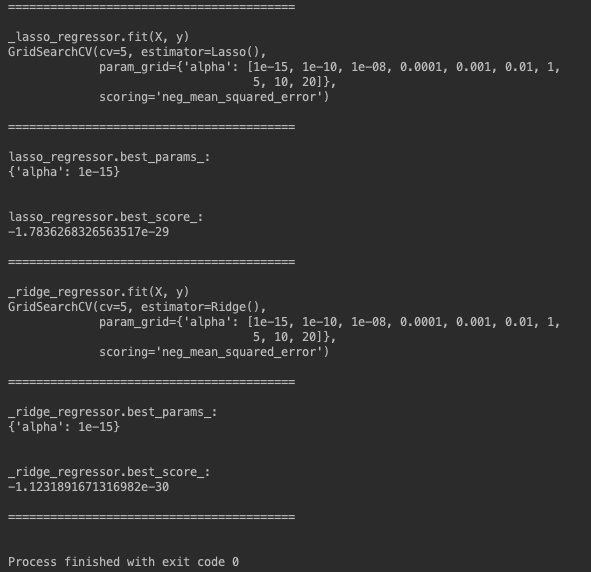
While output of alpha zero is correct:
fun: 2.1923913945084075e-24
message: 'Optimization terminated successfully.'
nfev: 632
nit: 12
status: 0
success: True
x: array([ 2.00000000e+00, -1.49737205e-13, -5.49916821e-13, 8.87767676e-13,
1.75335824e-13])
output of alpha non-zero has much higher loss and non of coefficients is zero as expected:
fun: 0.9714385008821652
message: 'Optimization terminated successfully.'
nfev: 527
nit: 6
status: 0
success: True
x: array([ 1.86644474e+00, 1.63986381e-02, 2.99944361e-03, 1.64568796e-12,
-6.72908469e-09])
Why coefficients of random features are not shrunk to zero and loss is so high?

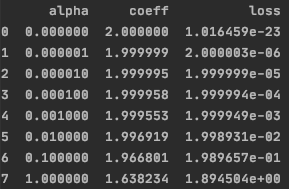
make_regression. – Famed