Running Jupyter Lab Version 3.0.11
I'd like to hide or remove cells, and/or hide cell input, and/or hide cell output in Jupyter Notebook and/or Jupyter Lab when converted to HTML and/or PDF. And, I want to choose which cells/input/output to remove and which to keep.
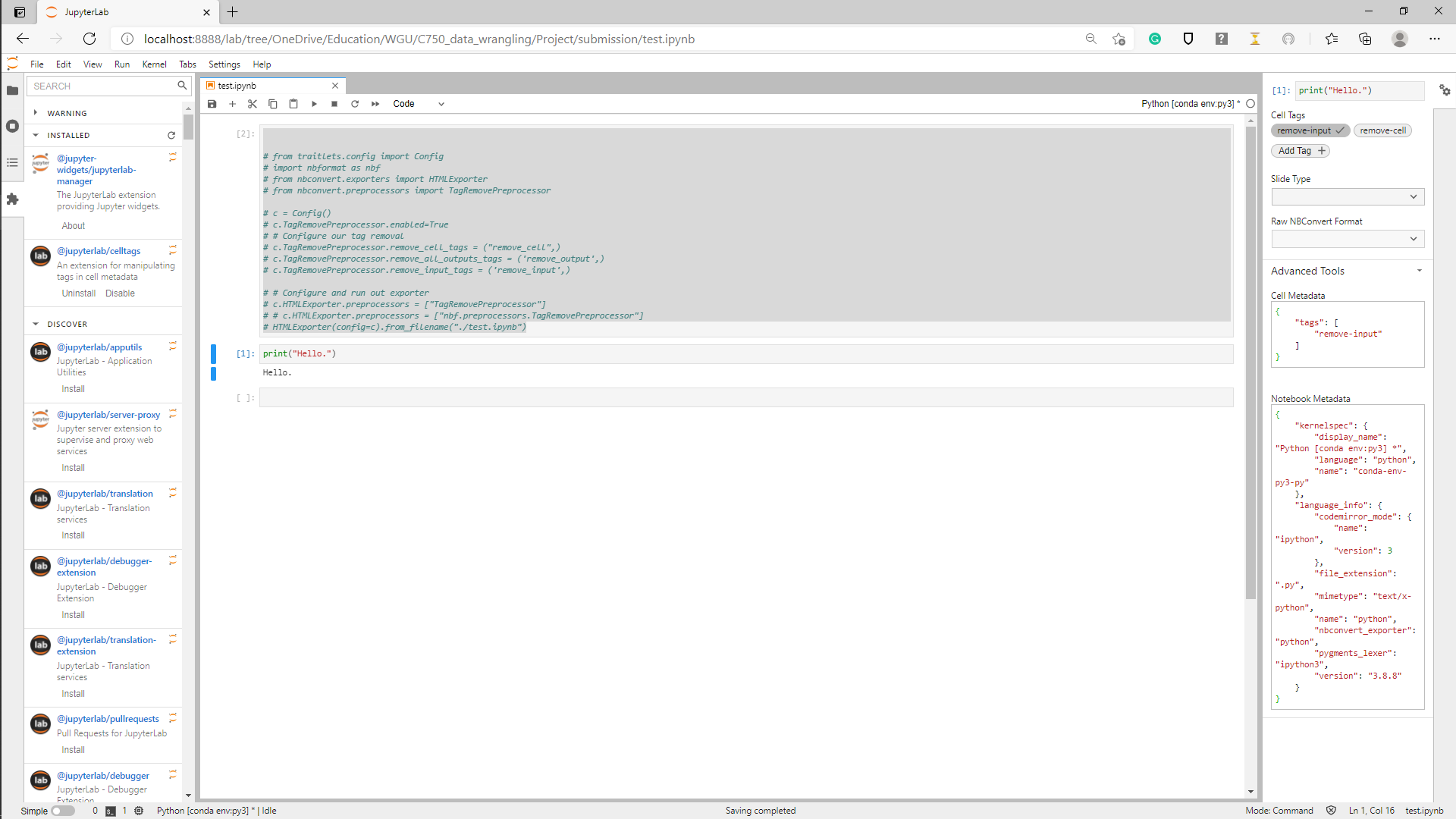
I'm trying Jupyter Lab's celltags extension, but it isn't working.
Here's the notebook with a "remove-input" cell tag (UPDATE: the celltags extension is no longer installed, but the results are the same):
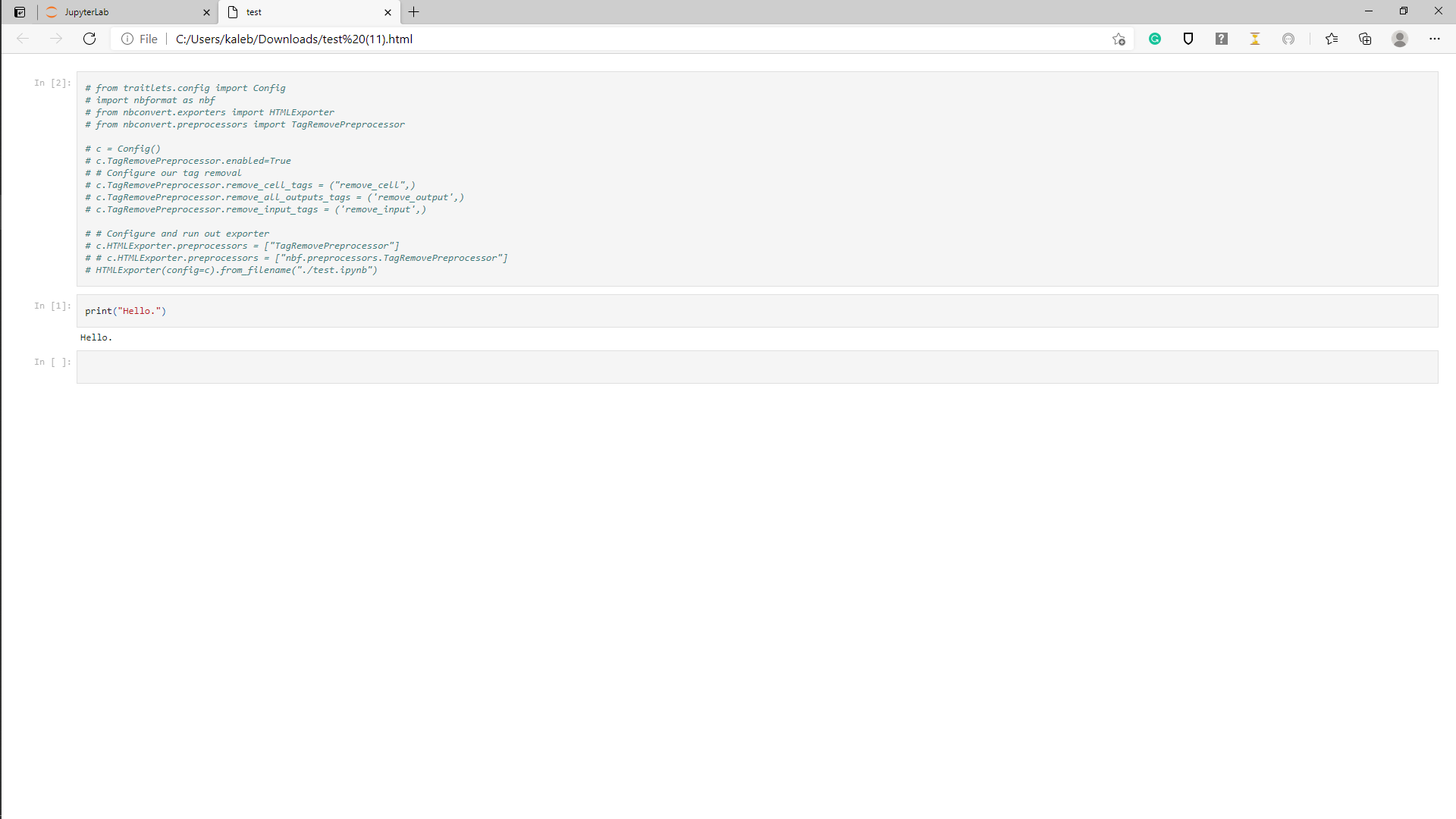
And, here's the HTML document it creates with the input not removed:
Hiding the cell in the notebook by clicking the tab next to it didn't translate to a hidden cell in the exported HTML doc.
As you can see, I have tried using TagRemovePreprocessor, but that just throws the error that the module is not found even though I have everything installed that I import. I tried a couple of fixes to that, which didn't work, but I don't really want to shave that yak if I don't need to.
I'd rather cell tags just work, like in R Studio markdown documents. At this point, it might even be easier to copy and paste my notebook to an RMD and run it with a Python kernel, since RMDs have that widget and easy tag built in and functional.
All that said, any Jupyter Notebook or Jupyter Lab solution is welcome, especially if it's just a simple toggle that actually works.
Thanks!
P.S. Here's the code from the commented out first cell, and the error from it:
from traitlets.config import Config
import nbformat as nbf
from nbconvert.exporters import HTMLExporter
from nbconvert.preprocessors import TagRemovePreprocessor
c = Config()
c.TagRemovePreprocessor.enabled=True
# Configure our tag removal
c.TagRemovePreprocessor.remove_cell_tags = ("remove_cell",)
c.TagRemovePreprocessor.remove_all_outputs_tags = ('remove_output',)
c.TagRemovePreprocessor.remove_input_tags = ('remove_input',)
# Configure and run out exporter
c.HTMLExporter.preprocessors = ["TagRemovePreprocessor"]
# c.HTMLExporter.preprocessors = ["nbf.preprocessors.TagRemovePreprocessor"]
HTMLExporter(config=c).from_filename("./test.ipynb")
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
<ipython-input-4-2a221250acbf> in <module>
14 c.HTMLExporter.preprocessors = ["TagRemovePreprocessor"]
15 # c.HTMLExporter.preprocessors = ["nbf.preprocessors.TagRemovePreprocessor"]
---> 16 HTMLExporter(config=c).from_filename("./test.ipynb")
~\anaconda3\envs\py3\lib\site-packages\nbconvert\exporters\templateexporter.py in __init__(self, config, **kw)
323 Template to use when exporting.
324 """
--> 325 super().__init__(config=config, **kw)
326
327 self.observe(self._invalidate_environment_cache,
~\anaconda3\envs\py3\lib\site-packages\nbconvert\exporters\exporter.py in __init__(self, config, **kw)
112 super().__init__(config=with_default_config, **kw)
113
--> 114 self._init_preprocessors()
115
116
~\anaconda3\envs\py3\lib\site-packages\nbconvert\exporters\templateexporter.py in _init_preprocessors(self)
488
489 def _init_preprocessors(self):
--> 490 super()._init_preprocessors()
491 conf = self._get_conf()
492 preprocessors = conf.get('preprocessors', {})
~\anaconda3\envs\py3\lib\site-packages\nbconvert\exporters\exporter.py in _init_preprocessors(self)
264 # Load user-specified preprocessors. Enable by default.
265 for preprocessor in self.preprocessors:
--> 266 self.register_preprocessor(preprocessor, enabled=True)
267
268
~\anaconda3\envs\py3\lib\site-packages\nbconvert\exporters\exporter.py in register_preprocessor(self, preprocessor, enabled)
225 # Preprocessor is a string, import the namespace and recursively call
226 # this register_preprocessor method
--> 227 preprocessor_cls = import_item(preprocessor)
228 return self.register_preprocessor(preprocessor_cls, enabled)
229
~\anaconda3\envs\py3\lib\site-packages\traitlets\utils\importstring.py in import_item(name)
36 else:
37 # called with un-dotted string
---> 38 return __import__(parts[0])
ModuleNotFoundError: No module named 'TagRemovePreprocessor'
I have installed nbconvert, and TagRemovePreprocessor doesn't throw an error in its import statement.
I can run c.HTMLExporter.preprocessors = ["nbf.preprocessors.TagRemovePreprocessor"] instead of c.HTMLExporter.preprocessors = ["TagRemovePreprocessor"], and I get the same error thrown from the same line except it says there is no module named nbf.

remove-inputwith a hyphen, but your code looks forremove_inputwith an underscore. I typically remove input cells using the command line interface with something like:jupyter nbconvert mynotebook.ipynb --TagRemovePreprocessor.enabled=True --TagRemovePreprocessor.remove_cell_tags remove_cell. More here: nbconvert.readthedocs.io/en/latest/removing_cells.html – Opinion