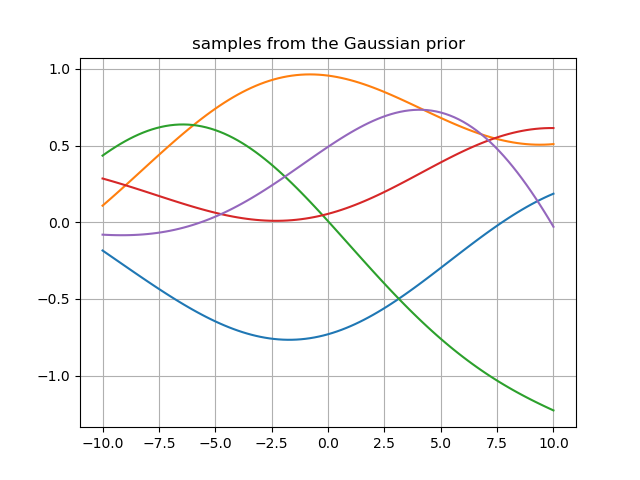
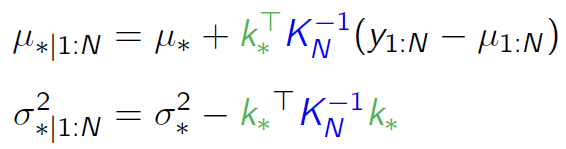I have created and sampled a jointly Gaussian prior with mean=0 using the code below:
import numpy as np
import matplotlib.pyplot as plt
from math import pi
from scipy.spatial.distance import cdist
import scipy.stats as sts
x_prior = np.linspace(-10,10,101)
x_prior = x_prior.reshape(-1,1)
mu = np.zeros(x_prior.shape)
#defining the Kernel for the covariance function
def sec(a,b, length_scale , sigma) :
K = sigma * np.exp(-1/(2*length_scale) * cdist(a,b)**2)
return K
#defining the Gaussian Process prior
def GP(a , b, mu , kernel , length_scale, sigma , samples ) :
f = np.random.multivariate_normal(mu.flatten(), kernel(a ,b , length_scale , sigma ) , samples)
return f
prior = GP(x_prior ,x_prior, mu , sec , 100, 1 , 5)
plt.figure()
plt.grid()
plt.title('samples from the Gaussian prior')
plt.plot(x_prior , prior.T)
plt.show()
Then, when adding in some 'observed' data, I wish to compute the posterior over these points but this is where I become stuck.
Here's my code for introducing new data:
x_train = np.array([-10,-8,5,-1,2])
x_train = x_train.reshape(-1,1)
def straight_line(m , x , c):
y = 5*x + c
return y
ytrain = straight_line(5 , x_train , 0)
It's my understanding that you calculate a conditional distribution over the new data given the prior and new x values associated with the observed data.
Do you then wish to update the multivariate prior to become the posterior by performing some sort of change to the mean values to include the new y values?
I have used the following resources to try and attempt this:
http://katbailey.github.io/post/gaussian-processes-for-dummies/ https://www.robots.ox.ac.uk/~mebden/reports/GPtutorial.pdf
but I'm really trying to understand what happens at each stage, and why, so that when I get a posterior (which I can't do) I know exactly how I got there.
Here's some solutions I've been trying to implement but so far no avail:
K_train = sec(x_train , x_train , 1,1)
K_prior = sec(x_prior , x_prior , 1,1)
K_pt = sec(x_prior , x_train , 1,1)
K_tp = sec(x_train , x_prior , 1,1) ## = k_tp transpose
prior = sts.multivariate_normal(mu.flatten(), K_prior)
#mean_test = np.dot(K_p , np.linalg.inv(K_prior))
mean_function = np.dot(np.dot(K_tp ,np.linalg.inv(K_prior).T) , prior )
covariance_function = K_train - np.dot(np.dot(K_tp ,np.linalg.inv(K_prior).T) , K_pt)