The short version of my problem is the following: I have a histogram of some data (density of planets) which seems to have 3 peeks. Now I want to fit 3 gaussians to this histogram.
I am expecting this outcome.

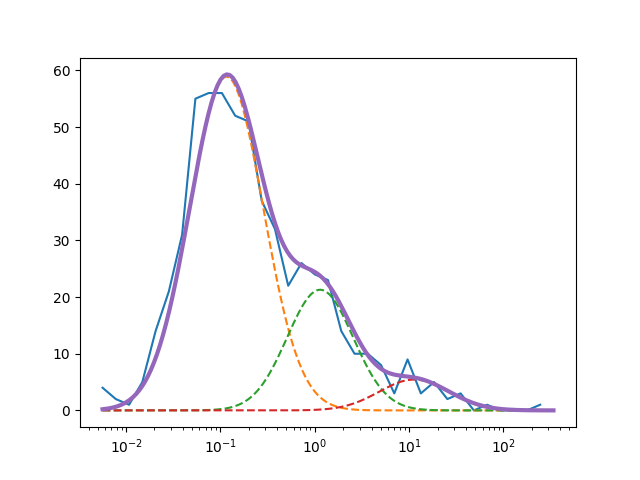
I used different methods to fit my gauss: curve_fit, least square and GaussianMixture from sklearn.mixture. With Curve_fit I get a pretty good fit

but it isn't good enough if you compare it to my expected outcome. With least square I get a "good fit"
 but my gaussians are nonsense, and with GaussianMixture I don't get anywhere, because I can't really adept the codes I've seen in Examples to my problem.
but my gaussians are nonsense, and with GaussianMixture I don't get anywhere, because I can't really adept the codes I've seen in Examples to my problem.
At this point I have three questions:
Most important: How can I get a better fit for my third gaussian? I already tried to adjust the initial values for p0, but then the gaussian becomes even worst or it doesn't find the parameters at all.
What is wrong with my least-square Code? Why does it give me such strange gaussians? And is there a way to fix this? My Guess: Is it because least-square does everything to minimize the error between fit and actual data?
How would I do the whole thing with GaussianMixture? I found this post
but couldn't adapt it to my problem.
I really want to understand how to make fits properly, since I will have to do them a lot in the future. The problem is that I am not very good in statistics and just started programming in python.
Here are my three different codes:
Curvefit
import numpy as np
import math
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
hist, bin_edges = np.histogram(Density, bins=np.logspace(np.log10(MIN), np.log10(MAX), 32))
bin_centres = (bin_edges[:-1] + bin_edges[1:])/2
# Define model function to be used to fit to the data above:
def triple_gaussian( x,*p ):
(c1, mu1, sigma1, c2, mu2, sigma2, c3, mu3, sigma3) = p
res = np.divide(1,x)*c1 * np.exp( - (np.log(x) - mu1)**2.0 / (2.0 * sigma1**2.0) ) \
+ np.divide(1,x)*c2 * np.exp( - (np.log(x) - mu2)**2.0 / (2.0 * sigma2**2.0) ) \
+ np.divide(1,x)*c3 * np.exp( - (np.log(x) - mu3)**2.0 / (2.0 * sigma3**2.0) )
return res
# p0 is the initial guess for the fitting coefficients (A, mu and sigma above)
p0 = [60., 1, 1., 30., 1., 1.,10., 1., 1]
coeff, var_matrix = curve_fit(triple_gaussian, bin_centres, hist, p0=p0)
# Get the fitted curve
hist_fit = triple_gaussian(bin_centres, *coeff)
c1 =coeff[0]
mu1 =coeff[1]
sigma1 =coeff[2]
c2 =coeff[3]
mu2 =coeff[4]
sigma2 =coeff[5]
c3 =coeff[6]
mu3 =coeff[7]
sigma3 =coeff[8]
x= bin_centres
gauss1= np.divide(1,x)*c1 * np.exp( - (np.log(x) - mu1)**2.0 / (2.0 * sigma1**2.0) )
gauss2= np.divide(1,x)*c2 * np.exp( - (np.log(x) - mu2)**2.0 / (2.0 * sigma2**2.0) )
gauss3= np.divide(1,x)*c3 * np.exp( - (np.log(x) - mu3)**2.0 / (2.0 * sigma3**2.0) )
plt.plot(x,gauss1, 'g',label='gauss1')
plt.plot(x,gauss2, 'b', label='gauss2')
plt.plot(x,gauss3, 'y', label='gauss3')
plt.gca().set_xscale("log")
plt.legend(loc='upper right')
plt.ylim([0,70])
plt.suptitle('Triple log Gaussian fit over all Data', fontsize=20)
plt.xlabel('log(Density)')
plt.ylabel('Number')
plt.hist(Density, bins=np.logspace(np.log10(MIN), np.log10(MAX), 32), label='all Densities')
plt.plot(bin_centres, hist, label='Test data')
plt.plot(bin_centres, hist_fit, label='Fitted data')
plt.gca().set_xscale("log")
plt.ylim([0,70])
plt.suptitle('triple log Gaussian fit using curve_fit', fontsize=20)
plt.xlabel('log(Density)')
plt.ylabel('Number')
plt.legend(loc='upper right')
plt.annotate(Text1, xy=(0.01, 0.95), xycoords='axes fraction')
plt.annotate(Text2, xy=(0.01, 0.90), xycoords='axes fraction')
plt.savefig('all Densities_gauss')
plt.show()
Leastsquare
The fit itselfe doesn't look to bad, but the 3 gaussians are horrible. See here

# I only have x-data, so to get according y-data I make my histogram and
#use the bins as x-data and the numbers (hist) as y-data.
#Density is a Dataset of 581 Values between 0 and 340.
hist, bin_edges = np.histogram(Density, bins=np.logspace(np.log10(MIN), np.log10(MAX), 32))
x = (bin_edges[:-1] + bin_edges[1:])/2
y = hist
#define tripple gaussian
def triple_gaussian( p,x ):
(c1, mu1, sigma1, c2, mu2, sigma2, c3, mu3, sigma3) = p
res = np.divide(1,x)*c1 * np.exp( - (np.log(x) - mu1)**2.0 / (2.0 * sigma1**2.0) ) \
+ np.divide(1,x)*c2 * np.exp( - (np.log(x) - mu2)**2.0 / (2.0 * sigma2**2.0) ) \
+ np.divide(1,x)*c3 * np.exp( - (np.log(x) - mu3)**2.0 / (2.0 * sigma3**2.0) )
return res
def errfunc(p,x,y):
return y-triple_gaussian(p,x)
p0=[]
p0 = [60., 0.1, 1., 30., 1., 1.,10., 10., 1.]
fit = optimize.leastsq(errfunc,p0,args=(x,y))
print('fit', fit)
plt.plot(x,y)
plt.plot(x,triple_gaussian(fit[0],x), 'r')
plt.gca().set_xscale("log")
plt.ylim([0,70])
plt.suptitle('Double log Gaussian fit over all Data', fontsize=20)
plt.xlabel('log(Density)')
plt.ylabel('Number')
c1, mu1, sigma1, c2, mu2, sigma2, c3, mu3, sigma3=fit[0]
print('c1', c1)
gauss1= np.divide(1,x)*c1 * np.exp( - (np.log(x) - mu1)**2.0 / (2.0 * sigma1**2.0) )
gauss2= np.divide(1,x)*c2 * np.exp( - (np.log(x) - mu2)**2.0 / (2.0 * sigma2**2.0) )
gauss3= np.divide(1,x)*c3 * np.exp( - (np.log(x) - mu3)**2.0 / (2.0 * sigma3**2.0) )
plt.plot(x,gauss1, 'g')
plt.plot(x,gauss2, 'b')
plt.plot(x,gauss3, 'y')
plt.gca().set_xscale("log")
plt.ylim([0,70])
plt.suptitle('Double log Gaussian fit over all Data', fontsize=20)
plt.xlabel('log(Density)')
plt.ylabel('Number')
GaussianMixture
As I said befor, I don't really understand GaussianMixture. I don't know if I have to define a triplegauss like before, or if it's enough to define the gauss, and GaussianMixture will find out that there is a triple gauss on its own. I also don't understand where I have to use which data, because when I use the bins and hist values, then the "fitted curve" are just the datapoints connected with each other. So I figured that I used the wrong data.
The parts I don't understand are #Fit GMM and #Construct function manually as sum of gaussians.
hist, bin_edges = np.histogram(Density, bins=np.logspace(np.log10(MIN), np.log10(MAX), 32))
bin_centres = (bin_edges[:-1] + bin_edges[1:])/2
plt.hist(Density, bins=np.logspace(np.log10(MIN), np.log10(MAX), 32), label='all Densities')
plt.gca().set_xscale("log")
plt.ylim([0,70])
# Define simple gaussian
def gauss_function(x, amp, x0, sigma):
return np.divide(1,x)*amp * np.exp(-(np.log(x) - x0) ** 2. / (2. * sigma ** 2.))
# My Data
samples = Density
# Fit GMM
gmm = GaussianMixture(n_components=3, covariance_type="full", tol=0.00001)
gmm = gmm.fit(X=np.expand_dims(samples, 1))
gmm_x= bin_centres
gmm_y= hist
# Construct function manually as sum of gaussians
gmm_y_sum = np.full_like(gmm_x, fill_value=0, dtype=np.float32)
for m, c, w in zip(gmm.means_.ravel(), gmm.covariances_.ravel(), gmm.weights_.ravel()):
gauss = gauss_function(x=gmm_x, amp=1, x0=m, sigma=np.sqrt(c))
gmm_y_sum += gauss / np.trapz(gauss, gmm_x) *w
# Make regular histogram
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=[8, 5])
ax.hist(samples, bins=np.logspace(np.log10(MIN), np.log10(MAX), 32), label='all Densities')
ax.plot(gmm_x, gmm_y, color="crimson", lw=4, label="GMM")
ax.plot(gmm_x, gmm_y_sum, color="black", lw=4, label="Gauss_sum", linestyle="dashed")
plt.gca().set_xscale("log")
plt.ylim([0,70])
# Annotate diagram
ax.set_ylabel("Probability density")
ax.set_xlabel("Arbitrary units")
# Make legend
plt.legend()
plt.show()
I hope that anyone can help me at least with one of my questions. As I said befor, if anything is missing or if you need more information please tell me.
Thanks in advance!
--Edit-- Here is my Data.

Densityvalues available by any chance? Right now it is not possible to test your code. – Lyricist